

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148398.1 + phase: 2 /pseudo/partial
(2332 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
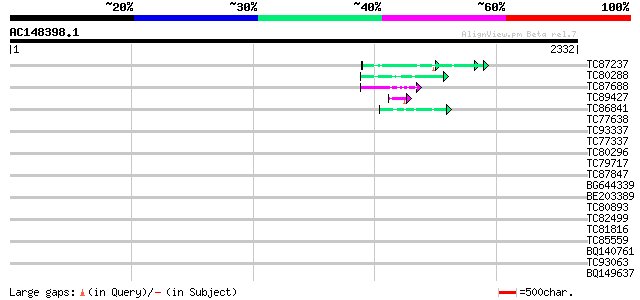
Score E
Sequences producing significant alignments: (bits) Value
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 78 4e-14
TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein... 65 2e-10
TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~un... 50 1e-05
TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed pr... 46 2e-04
TC86841 similar to PIR|B86416|B86416 unknown protein 16040-1118... 44 6e-04
TC77638 similar to GP|20161455|dbj|BAB90379. ankyrin-like protei... 42 0.002
TC93337 weakly similar to GP|21240660|gb|AAM44371.1 hypothetical... 42 0.003
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 42 0.004
TC80296 similar to PIR|H86265|H86265 protein F3F19.18 [imported]... 41 0.005
TC79717 similar to GP|10086499|gb|AAG12559.1 Unknown Protein {Ar... 41 0.005
TC87847 similar to GP|15929245|gb|AAH15068.1 Similar to adaptor-... 41 0.005
BG644339 weakly similar to PIR|T48401|T484 histone deacetylase-l... 41 0.006
BE203389 weakly similar to PIR|T06754|T0 DNA-directed RNA polyme... 40 0.008
TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {... 40 0.014
TC82499 SP|Q9XHM1|IF38_MEDTR Eukaryotic translation initiation f... 39 0.018
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 39 0.018
TC85559 similar to PIR|T07012|T07012 acetyl-CoA carboxylase (EC ... 39 0.018
BQ140761 weakly similar to GP|18076245|emb phosphophoryn {Rattus... 39 0.018
TC93063 similar to PIR|T08852|T08852 lustrin A - California red ... 39 0.024
BQ149637 similar to PIR|D85135|D85 hypothetical protein AT4g1261... 33 0.99
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 78.2 bits (191), Expect = 4e-14
Identities = 106/520 (20%), Positives = 202/520 (38%), Gaps = 36/520 (6%)
Frame = +1
Query: 1448 QKKEEQEESKEVPGNDNTGIEMDQDFQADAVSLSEDSSENEDCDGENEELESEMGPTGPD 1507
+ K+E EE+ E D E +Q+ S+ ++ GEN E +S+ G +
Sbjct: 343 KSKQENEETSETNSKDKENEESNQN-----------GSDAKEQVGENHEQDSKQGTEETN 489
Query: 1508 SEAVGEKIWDQNEDETPDDTGEKYESGPSVKDGDGSNKELRAKDDSTSDQSGDDSCDEGD 1567
GEK + D+ +DT S V+DG+ +N+ A++++ S + + +
Sbjct: 490 GTEGGEK---EEHDKIKEDT----SSDNQVQDGEKNNE---AREENYSGDNASSAVVDNK 639
Query: 1568 AQNDEAAAQNEFDDEENGD-DVNMDKEAAHSDATGLKPDEPDHSSDMEIDLNANEDVDPI 1626
+Q + +FD +E + ++ K + +E S+D I N+ +
Sbjct: 640 SQESSNKTEEQFDKKEKNEFELESQKNS----------NETTESTDSTITQNSQGNESEK 789
Query: 1627 EEGDQEGHDDSAENGNQEDETCPPDEIMEEAHTEVDVSSEKDDLGQEHQENDDMNSMDPK 1686
++ E +D + ++ DE E +V + G + E + S D
Sbjct: 790 DQAQTE-NDTPKGSASESDEQKQEQEQNNTTKDDVQTTDTSSQNGNDTTEKQNETSED-A 963
Query: 1687 NDTSESSDVVNPQVSNVDLASQSKSDLQTSGSENIASQSNLSNSHH------DFGNPAVT 1740
N E S +N +N D S D S + +S++ N++H NP
Sbjct: 964 NSKKEDSSALNTTPNNEDSKSGVAGDQADSTTTTSSSETQDGNTNHGEYKDTTNENPEKN 1143
Query: 1741 GG-------------FPSSDMSEMDINMSDSSNTGG------FSKTQPKSHLPQHEHSFS 1781
G F + D + + ++ +S+T S + KS Q++++ S
Sbjct: 1144 SGQEGTQESGSSSNTFDNKDAASNKVQLTTTSDTSSEQKKDESSSAESKSESSQNDNANS 1323
Query: 1782 QEKQTNPSRSTGDALDFRKEKINVSGDLPEDNIEHHGEMDDNNADEYGYVSEFEKGTTQA 1841
+ T S D D ++ S +N +NN+DE S+ + T +
Sbjct: 1324 GQSNTTSDESANDNKD--SSQVTTS----SENSAEGNSNTENNSDENQNDSKNNENTNDS 1485
Query: 1842 LGPATLEQVDRNIDVDKVDTESRAGE----DANLQLEKQNSE-----IDSVSNSSLLPKN 1892
+ V+ N + + T++ E + +++ +K+N+E +D+ SNS+
Sbjct: 1486 GNTSNDANVNENQNENAAQTKTSENEGDAQNESVESKKENNESAHKDVDNNSNSN----- 1650
Query: 1893 EKRDQANMPVMEKSQDDGSVKPLGS-ANIDPESHLEDLIS 1931
DQ + D S LG+ + ESH D+ S
Sbjct: 1651 ---DQGSSDTSVTQDDKESRVDLGTLPESNGESHHNDVSS 1761
Score = 74.3 bits (181), Expect = 5e-13
Identities = 104/537 (19%), Positives = 212/537 (39%), Gaps = 18/537 (3%)
Frame = +1
Query: 1452 EQEESKEVPGNDNTGIEMDQDFQADAVSLSEDSSENEDCD---GENEELESEMGPTGPDS 1508
E EESKE + ++ + D +++ E +D + ENEE + E
Sbjct: 73 ENEESKETGESSEEKSHLENEENKDEEKSKQENEEIKDGEKIQQENEENKDE-------- 228
Query: 1509 EAVGEKIWDQNEDETPDDTGEKYESGPSVKDGDGSNKELRAKDDSTSDQSGDDSCDEG-- 1566
EK +NE+ + EK + +K +G KE + S Q +++ +
Sbjct: 229 ----EKSQQENEENKDE---EKSQQENELKKNEGGEKETGEITEEKSKQENEETSETNSK 387
Query: 1567 DAQNDEAAAQNEFDDEENGDDVNMDKEAAHSDATGLKPDEPDHSSDMEIDLNANEDVDPI 1626
D +N+E+ E+ G++ D + + G + E + ++ D +++ V
Sbjct: 388 DKENEESNQNGSDAKEQVGENHEQDSKQGTEETNGTEGGEKEEHDKIKEDTSSDNQVQDG 567
Query: 1627 EEGDQEGHDD-SAENGNQEDETCPPDEIMEEAHTEVDVSSEKDDLGQEHQENDDMNSMDP 1685
E+ ++ ++ S +N + E + + D EK++ E Q+N
Sbjct: 568 EKNNEAREENYSGDNASSAVVDNKSQESSNKTEEQFD-KKEKNEFELESQKN-------- 720
Query: 1686 KNDTSESSDVVNPQVSNVDLASQSKSDLQTSGSENIASQSNLSNSHHDFGNPAVTGGFPS 1745
N+T+ES+D Q S + + + ++ + + AS+S+ + N
Sbjct: 721 SNETTESTDSTITQNSQGNESEKDQAQTENDTPKGSASESDEQKQEQEQNN--------- 873
Query: 1746 SDMSEMDINMSDSSNTGGFSKTQPKSHLPQHEHSFSQEKQ------TNPSRSTGDALDFR 1799
++ D+ +D+S+ G T+ ++ + +S ++ N +G A D
Sbjct: 874 --TTKDDVQTTDTSSQNGNDTTEKQNETSEDANSKKEDSSALNTTPNNEDSKSGVAGDQA 1047
Query: 1800 KEKINVSGDLPEDNIEHHGEMDDNNADEYGYVSEFEKGTTQALGPATLEQVDRNIDVDKV 1859
S +D +HGE D +E + ++G TQ G ++ +++ +KV
Sbjct: 1048 DSTTTTSSSETQDGNTNHGEYKD-TTNENPEKNSGQEG-TQESGSSSNTFDNKDAASNKV 1221
Query: 1860 ------DTESRAGEDANLQLEKQNSEIDSVSNSSLLPKNEKRDQANMPVMEKSQDDGSVK 1913
DT S +D + E + SE N++ N D++ + SQ S +
Sbjct: 1222 QLTTTSDTSSEQKKDESSSAESK-SESSQNDNANSGQSNTTSDESANDNKDSSQVTTSSE 1398
Query: 1914 PLGSANIDPESHLEDLISISRSYLGENTHKLSQLSVNDEELGKYHEPCDAPDHVKDN 1970
N + E++ ++ + S++ ENT+ S ND + + A +N
Sbjct: 1399 NSAEGNSNTENNSDENQNDSKN--NENTNDSGNTS-NDANVNENQNENAAQTKTSEN 1560
Score = 63.5 bits (153), Expect = 9e-10
Identities = 68/348 (19%), Positives = 134/348 (37%), Gaps = 22/348 (6%)
Frame = +1
Query: 1447 MQKKEEQEESKEVPGNDNTGIEMDQDFQADAVSLSEDSSENEDCDGENEELESEMGPTGP 1506
++ ++ E+ E + T + + D D+ + + + ++ E E T
Sbjct: 703 LESQKNSNETTESTDSTITQNSQGNESEKDQAQTENDTPKGSASESDEQKQEQEQNNTTK 882
Query: 1507 DSEAVGEKIWDQNEDETPDDTGEKYESGPSVKDGDGSNKELRAKDDSTSDQSGDDSCDEG 1566
D + QN ++T + E E S K+ + +DS S +GD +
Sbjct: 883 DDVQTTDTS-SQNGNDTTEKQNETSEDANSKKEDSSALNTTPNNEDSKSGVAGDQADSTT 1059
Query: 1567 DAQNDEAAAQN----EFDDEENGD-DVNMDKEAAHSDATGLKPDEPDHSSDMEIDLNANE 1621
+ E N E+ D N + + N +E + + ++ ++ L
Sbjct: 1060 TTSSSETQDGNTNHGEYKDTTNENPEKNSGQEGTQESGSSSNTFDNKDAASNKVQLTTTS 1239
Query: 1622 DVDPIEEGDQ----EGHDDSAENGN----QEDETCPPDEIMEEAHTEVDVSSEKDDLGQE 1673
D ++ D+ E +S++N N Q + T + ++V SSE G
Sbjct: 1240 DTSSEQKKDESSSAESKSESSQNDNANSGQSNTTSDESANDNKDSSQVTTSSENSAEGNS 1419
Query: 1674 HQEND-DMNSMDPKN-----DTSESSDVVNPQVSNVDLASQSKSDLQTSGSENIASQSNL 1727
+ EN+ D N D KN D+ +S+ N + + A+Q+K+ ++N + +S
Sbjct: 1420 NTENNSDENQNDSKNNENTNDSGNTSNDANVNENQNENAAQTKTSENEGDAQNESVESKK 1599
Query: 1728 SNS---HHDFGNPAVTGGFPSSDMSEMDINMSDSSNTGGFSKTQPKSH 1772
N+ H D N + + SSD S + + G ++ +SH
Sbjct: 1600 ENNESAHKDVDNNSNSNDQGSSDTSVTQDDKESRVDLGTLPESNGESH 1743
>TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein T28I19.100 -
Arabidopsis thaliana, partial (14%)
Length = 1460
Score = 65.5 bits (158), Expect = 2e-10
Identities = 78/365 (21%), Positives = 145/365 (39%), Gaps = 3/365 (0%)
Frame = +3
Query: 1442 CLLSMMQKKEEQEESKEVPGNDNT---GIEMDQDFQADAVSLSEDSSENEDCDGENEELE 1498
C + Q K ++ KE ND E DQ + L E + +G EE E
Sbjct: 237 CFWLIYQVKHNHDKKKEFDKNDTKLPIRTETDQILKLGRKDLHPGKVEADKNEGHEEEEE 416
Query: 1499 SEMGPTGPDSEAVGEKIWDQNEDETPDDTGEKYESGPSVKDGDGSNKELRAKDDSTSDQS 1558
E + + ++ E + GE+ + ++ + + E R + D ++
Sbjct: 417 DEH---------IVYNMQNKREHDEQQQEGEEGNKHETEEESEDNVHERREEQDEEENKH 569
Query: 1559 GDDSCDEGDAQNDEAAAQNEFDDEENGDDVNMDKEAAHSDATGLKPDEPDHSSDMEIDLN 1618
G + +E +++++E E+ G DV +D E DH +
Sbjct: 570 GAEVQEENESKSEEV--------EDEGGDVEID--------------ENDHEKSEADNDR 683
Query: 1619 ANEDVDPIEEGDQEGHDDSAENGNQEDETCPPDEIMEEAHTEVDVSSEKDDLGQEHQEND 1678
+E VD ++ ++EG DD EN ++EDE ++E + E + +EH + D
Sbjct: 684 EDEVVDEEKDKEEEG-DDETENEDKEDE--EKGGLVE--------NHENHEAREEHYKAD 830
Query: 1679 DMNSMDPKNDTSESSDVVNPQVSNVDLASQSKSDLQTSGSENIASQSNLSNSHHDFGNPA 1738
D +S + S++ N + S++ L + K + +T+ S+ N+S+ G
Sbjct: 831 DASSAVAHDTHETSTETGNLEHSDLSLQNTRKPENETNHSDESYGSQNVSDLKVTEGE-- 1004
Query: 1739 VTGGFPSSDMSEMDINMSDSSNTGGFSKTQPKSHLPQHEHSFSQEKQTNPSRSTGDALDF 1798
+T G S+ + + SN +K + L + + E +N S TGD
Sbjct: 1005 LTDGVSSNATAGKETGNDSFSNETAKTKPDSQLDLSSNLTAVITEASSN-SSGTGDDTSS 1181
Query: 1799 RKEKI 1803
E+I
Sbjct: 1182 SSEQI 1196
>TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~unknown protein
{Arabidopsis thaliana}, partial (46%)
Length = 2077
Score = 49.7 bits (117), Expect = 1e-05
Identities = 58/252 (23%), Positives = 105/252 (41%), Gaps = 1/252 (0%)
Frame = +2
Query: 1444 LSMMQKKEEQEESKEVPGNDNTGIEMDQDFQADAVSLSEDSSENEDCDGENEELESEMGP 1503
L+ K E+ +VP + +E + + + + E +D D + E +E +
Sbjct: 278 LNQFVKNPEEIAQFKVPLDVGKKLEKKKRVELEEEESDDFDEELDDDDDDFEGVEKKKAK 457
Query: 1504 TGPDSEAVGEKIWDQNEDETPDDTGEKYESGPSVKDGDGSNKELRAKDDSTSDQSGDDSC 1563
G + E E+ D++E+ + D+ E+ E VK G +K L+ + + + +D+
Sbjct: 458 GGSEGEDDFEEEDDEDEEGSEDEDDEEDEK-EKVKGGGIEDKFLKIDELTEYLEKEEDNY 634
Query: 1564 DEGDAQNDEAAAQNEFDDE-ENGDDVNMDKEAAHSDATGLKPDEPDHSSDMEIDLNANED 1622
++G+ + DEA +E DDE E + MD E D+ D D E ED
Sbjct: 635 EKGE-ERDEADEDSEEDDELEKAGEFEMDDE----------DDDDDDEEDEEA-----ED 766
Query: 1623 VDPIEEGDQEGHDDSAENGNQEDETCPPDEIMEEAHTEVDVSSEKDDLGQEHQENDDMNS 1682
+ + D G G E + D+++E + E D+ S K Q+ +
Sbjct: 767 MGNVRYEDFFG-------GKNEKGSKRKDQLLEVSGDEDDMESTK----QKKRTASTYEK 913
Query: 1683 MDPKNDTSESSD 1694
KN +S+D
Sbjct: 914 QRGKNSIKDSAD 949
>TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed protein
{Arabidopsis thaliana}, partial (50%)
Length = 753
Score = 45.8 bits (107), Expect = 2e-04
Identities = 29/103 (28%), Positives = 49/103 (47%), Gaps = 9/103 (8%)
Frame = +1
Query: 1559 GDDSCDEGDAQNDEAAAQNEFDDEENGDDVNMDKEAAHSDATGLKPDEPDHSSDMEIDLN 1618
G+ +C+E N + + + DDE++ DDVN + + D +G DE D +D E D
Sbjct: 268 GNHTCEE----NKDGSETEDDDDEDDDDDVNDEDDDNDEDFSG---DEDDEDADPEDDPV 426
Query: 1619 AN---------EDVDPIEEGDQEGHDDSAENGNQEDETCPPDE 1652
N ED D ++ + +G D+ + +DE PP +
Sbjct: 427 PNGAGGSDDDDEDDDDDDDDNDDGEDEDEDEEEDDDEDQPPSK 555
Score = 38.9 bits (89), Expect = 0.024
Identities = 30/107 (28%), Positives = 49/107 (45%), Gaps = 5/107 (4%)
Frame = +1
Query: 1507 DSEAVGEKIWDQNED--ETPDDTGEKYESGPSVKDGDGSNKELRAKDDSTSDQSGDDSCD 1564
D + G ++N+D ET DD E D D N E DD+ D SGD+ +
Sbjct: 253 DQLSKGNHTCEENKDGSETEDDDDE--------DDDDDVNDE---DDDNDEDFSGDEDDE 399
Query: 1565 EGDAQND---EAAAQNEFDDEENGDDVNMDKEAAHSDATGLKPDEPD 1608
+ D ++D A ++ DDE++ DD + + + D + D+ D
Sbjct: 400 DADPEDDPVPNGAGGSDDDDEDDDDDDDDNDDGEDEDEDEEEDDDED 540
Score = 33.9 bits (76), Expect = 0.76
Identities = 26/95 (27%), Positives = 41/95 (42%)
Frame = +1
Query: 1538 KDGDGSNKELRAKDDSTSDQSGDDSCDEGDAQNDEAAAQNEFDDEENGDDVNMDKEAAHS 1597
KDG + + DD + DD NDE + +E D++ + +D + A S
Sbjct: 292 KDGSETEDDDDEDDDDDVNDEDDD--------NDEDFSGDEDDEDADPEDDPVPNGAGGS 447
Query: 1598 DATGLKPDEPDHSSDMEIDLNANEDVDPIEEGDQE 1632
D DE D D + D +ED D E+ D++
Sbjct: 448 D----DDDEDDDDDDDDNDDGEDEDEDEEEDDDED 540
>TC86841 similar to PIR|B86416|B86416 unknown protein 16040-11188 [imported]
- Arabidopsis thaliana, partial (17%)
Length = 2361
Score = 44.3 bits (103), Expect = 6e-04
Identities = 71/321 (22%), Positives = 127/321 (39%), Gaps = 26/321 (8%)
Frame = +2
Query: 1520 EDETPDDTGEKYESGPSVKD---GDGSNKELRAKDDSTSDQSGDDSCDEGDAQNDEAAAQ 1576
EDE PDD G + D G S + D S QS D+ +A+N ++
Sbjct: 443 EDENPDDVGSASIPAKIISDDNPGTASGYDDNLYKDINSYQSHRHPFDDNEAENGVSSVA 622
Query: 1577 NEFDDEENGDDVNMDKEAAHSDATGLKPDEPDHSSDM---EIDLNANEDVDPIEEGDQEG 1633
N+++ H+D G++P+E D+S + + L+ E + G
Sbjct: 623 T-----------NLEQLNLHTDEQGIEPEE-DNSDVLIPNHLQLHTPECFS-LSFGSFGP 763
Query: 1634 HDDSAENGNQEDETCPPDEIMEEAHTEVDVSSEKDDLGQEHQENDDMNSMDPKNDTSESS 1693
++ +G + P +EE DVS+ +G +N D + TS+ +
Sbjct: 764 KQNATVSGAGTHSSRPLQSNLEETSGATDVSA----IGSSDVKNPDYYGDEHIATTSDGN 931
Query: 1694 DVVNPQVSNVDLASQSKSD------LQTSGSENI-ASQSNLSNSHHDF-----GNPAVTG 1741
++ ++ VD + S L++ SE +Q + +S H+F P V
Sbjct: 932 NI--SHITGVDARTYEHSSISQPEALKSEPSETAQENQYSFPSSSHEFTYENAQQPEV-- 1099
Query: 1742 GFPSSDMSEMDINMSDSSNTGGFSKTQPKSHL------PQHEHSFS--QEKQTNPSRSTG 1793
+P S S N+S S+ ++ + P + L P+ + +S Q P++ +
Sbjct: 1100 AYPHSQTSSQIQNLSPFSSVMAYTNSLPNALLASTVQTPREDIPYSPFPVTQAMPAKYSN 1279
Query: 1794 DALDFRKEKINVSGDLPEDNI 1814
A IN+S L +NI
Sbjct: 1280 MASSIGGSTINMSEALRANNI 1342
>TC77638 similar to GP|20161455|dbj|BAB90379. ankyrin-like protein {Oryza
sativa (japonica cultivar-group)}, partial (62%)
Length = 2345
Score = 42.4 bits (98), Expect = 0.002
Identities = 46/204 (22%), Positives = 89/204 (43%), Gaps = 7/204 (3%)
Frame = +1
Query: 1569 QNDEAAAQNEFDDEENGDDVNMDKEAAHSDATGLKPDEPDHSSDMEIDLNANEDVDPIEE 1628
QN++ +++ + +E + E +S+A + + D D + +V E+
Sbjct: 436 QNEDVPQESKSEVKEQTEVREQVSETDNSNARQFEDNPGDLPEDAT---KGDSNVSSEEK 606
Query: 1629 GDQEGHDDSAENGNQEDETCPPDEIMEEAHTEVDVSSEK-------DDLGQEHQENDDMN 1681
++ + S+E+ EDE ++ E ++TE + E+ D ++ E+++ N
Sbjct: 607 SEENSTEKSSEDTKTEDEGKKTED--EGSNTENNKDGEEASTKESESDESEKKDESEENN 780
Query: 1682 SMDPKNDTSESSDVVNPQVSNVDLASQSKSDLQTSGSENIASQSNLSNSHHDFGNPAVTG 1741
D +SSD SNV+ + Q S+ AS+ N ++ D + V
Sbjct: 781 KSDSDESEKKSSDSNETTDSNVE---EKVEQSQNKESDENASEKNTDDNAKDQSSNEV-- 945
Query: 1742 GFPSSDMSEMDINMSDSSNTGGFS 1765
FPS SE+ +N ++ TG FS
Sbjct: 946 -FPSGAQSEL-LN-ETTTQTGSFS 1008
Score = 32.0 bits (71), Expect = 2.9
Identities = 33/177 (18%), Positives = 68/177 (37%)
Frame = +1
Query: 1651 DEIMEEAHTEVDVSSEKDDLGQEHQENDDMNSMDPKNDTSESSDVVNPQVSNVDLASQSK 1710
+++ +E+ +EV E+ ++ ++ E D+ N+ +++ + + SNV +S+
Sbjct: 442 EDVPQESKSEV---KEQTEVREQVSETDNSNARQFEDNPGDLPEDATKGDSNVSSEEKSE 612
Query: 1711 SDLQTSGSENIASQSNLSNSHHDFGNPAVTGGFPSSDMSEMDINMSDSSNTGGFSKTQPK 1770
+ SE+ ++ + + N + E + SD S S+ K
Sbjct: 613 ENSTEKSSEDTKTEDEGKKTEDEGSNTENNKDGEEASTKESE---SDESEKKDESEENNK 783
Query: 1771 SHLPQHEHSFSQEKQTNPSRSTGDALDFRKEKINVSGDLPEDNIEHHGEMDDNNADE 1827
S + E S +T S +EK+ S + D DDN D+
Sbjct: 784 SDSDESEKKSSDSNETTDSNV--------EEKVEQSQNKESDENASEKNTDDNAKDQ 930
Score = 31.2 bits (69), Expect = 4.9
Identities = 33/145 (22%), Positives = 60/145 (40%)
Frame = +1
Query: 1807 GDLPEDNIEHHGEMDDNNADEYGYVSEFEKGTTQALGPATLEQVDRNIDVDKVDTESRAG 1866
GDLPED D+N VS EK + +++ + K + E +
Sbjct: 550 GDLPED-----ATKGDSN------VSSEEKSEENS--------TEKSSEDTKTEDEGKKT 672
Query: 1867 EDANLQLEKQNSEIDSVSNSSLLPKNEKRDQANMPVMEKSQDDGSVKPLGSANIDPESHL 1926
ED E ++ + S ++EK+D++ KS D S K +N +S++
Sbjct: 673 EDEGSNTENNKDGEEASTKESESDESEKKDESE--ENNKSDSDESEKKSSDSNETTDSNV 846
Query: 1927 EDLISISRSYLGENTHKLSQLSVND 1951
E+ + +S E+ S+ + +D
Sbjct: 847 EE--KVEQSQNKESDENASEKNTDD 915
>TC93337 weakly similar to GP|21240660|gb|AAM44371.1 hypothetical protein
{Dictyostelium discoideum}, partial (7%)
Length = 743
Score = 42.0 bits (97), Expect = 0.003
Identities = 42/184 (22%), Positives = 78/184 (41%), Gaps = 19/184 (10%)
Frame = +1
Query: 1565 EGDAQNDEAAAQNEFDDEENGDDVNMDKEAAHSDATGLKPDEPDHSSDMEIDLNANEDVD 1624
E + DE + + ++E+G+ D E +D KP+ D S + +E +
Sbjct: 4 ETKVKEDEESRTRDGGEKEDGERGGEDDEMDENDPE--KPEVVDRSEEF-----VDEGKE 162
Query: 1625 PIEEGDQEGHDDSAENGNQ------------EDETCPPDEIMEEAHTEVDVSSEKDDLGQ 1672
EEGD++ +++S + GNQ E++ D H S+E + +
Sbjct: 163 KEEEGDEKENENSKDEGNQGLVESNNIHEAREEQYKGDDASSAVTHDTRTTSTETETVSM 342
Query: 1673 EHQE-NDDMNSMDPKNDTSESSDVVNPQ------VSNVDLASQSKSDLQTSGSENIASQS 1725
E+ + N +M P+N + + D + Q ++ V L + S+ S S N +
Sbjct: 343 ENSDVNAEMGITKPENKPNYTEDGIRSQPGSNFSITEVKLVVGAYSN---SSSSNETGNN 513
Query: 1726 NLSN 1729
+LSN
Sbjct: 514 SLSN 525
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 41.6 bits (96), Expect = 0.004
Identities = 29/123 (23%), Positives = 51/123 (40%), Gaps = 9/123 (7%)
Frame = +3
Query: 1518 QNEDETPDDTGEKYESGPSVKDGDGSNKELRAKDDSTSDQSGDDSCDEGDAQNDEAAAQN 1577
Q++D+ D GE +D DG +D+ +S+ G + + N + A +
Sbjct: 387 QSQDKQHDAEGEDGNDDEDEEDDDGDGAFGEGEDELSSEDGGGYGNNSNNKSNSKKAPEG 566
Query: 1578 ---------EFDDEENGDDVNMDKEAAHSDATGLKPDEPDHSSDMEIDLNANEDVDPIEE 1628
E +D+E+GDD + D + D + E D ++ + N E+ D EE
Sbjct: 567 GAGGADENGEEEDDEDGDDQDEDDDEDEDDDDEEEGGEEDEEEGVDEEDNEEEEEDEDEE 746
Query: 1629 GDQ 1631
Q
Sbjct: 747 ALQ 755
Score = 40.4 bits (93), Expect = 0.008
Identities = 31/134 (23%), Positives = 58/134 (43%)
Frame = +3
Query: 1481 SEDSSENEDCDGENEELESEMGPTGPDSEAVGEKIWDQNEDETPDDTGEKYESGPSVKDG 1540
S+ S+++ D E E+ + D G+ + + EDE + G Y G
Sbjct: 378 SKTQSQDKQHDAEGEDGNDDEDEEDDD----GDGAFGEGEDELSSEDGGGY--------G 521
Query: 1541 DGSNKELRAKDDSTSDQSGDDSCDEGDAQNDEAAAQNEFDDEENGDDVNMDKEAAHSDAT 1600
+ SN + +K G D + G+ ++DE + DD+E+ DD + ++ +
Sbjct: 522 NNSNNKSNSKKAPEGGAGGAD--ENGEEEDDEDGDDQDEDDDEDEDDDDEEEGGEEDEEE 695
Query: 1601 GLKPDEPDHSSDME 1614
G+ DE D+ + E
Sbjct: 696 GV--DEEDNEEEEE 731
Score = 38.9 bits (89), Expect = 0.024
Identities = 35/151 (23%), Positives = 60/151 (39%), Gaps = 5/151 (3%)
Frame = +3
Query: 1449 KKEEQEESKEVPGNDNTGIEMDQDFQAD-AVSLSEDSSENEDCDG----ENEELESEMGP 1503
K + Q++ + G D E ++D D A ED +ED G N + S+ P
Sbjct: 381 KTQSQDKQHDAEGEDGNDDEDEEDDDGDGAFGEGEDELSSEDGGGYGNNSNNKSNSKKAP 560
Query: 1504 TGPDSEAVGEKIWDQNEDETPDDTGEKYESGPSVKDGDGSNKELRAKDDSTSDQSGDDSC 1563
G A ++ ++ +DE DD +D+ + DD
Sbjct: 561 EGGAGGA--DENGEEEDDEDGDD-----------------------QDEDDDEDEDDDDE 665
Query: 1564 DEGDAQNDEAAAQNEFDDEENGDDVNMDKEA 1594
+EG +++E E ++EE D+ D+EA
Sbjct: 666 EEGGEEDEEEGVDEEDNEEEEEDE---DEEA 749
Score = 38.1 bits (87), Expect = 0.040
Identities = 37/136 (27%), Positives = 54/136 (39%)
Frame = +3
Query: 1559 GDDSCDEGDAQNDEAAAQNEFDDEENGDDVNMDKEAAHSDATGLKPDEPDHSSDMEIDLN 1618
GD D Q+D A ++ DDE+ DD D + A + E +
Sbjct: 372 GDSKTQSQDKQHD-AEGEDGNDDEDEEDD---DGDGAFGEGEDELSSEDGGGYGNNSNNK 539
Query: 1619 ANEDVDPIEEGDQEGHDDSAENGNQEDETCPPDEIMEEAHTEVDVSSEKDDLGQEHQEND 1678
+N P EG G D ENG +ED+ D+ E D E DD +E E D
Sbjct: 540 SNSKKAP--EGGAGGAD---ENGEEEDDEDGDDQ------DEDDDEDEDDDDEEEGGEED 686
Query: 1679 DMNSMDPKNDTSESSD 1694
+ +D +++ E D
Sbjct: 687 EEEGVDEEDNEEEEED 734
>TC80296 similar to PIR|H86265|H86265 protein F3F19.18 [imported] -
Arabidopsis thaliana, partial (34%)
Length = 1734
Score = 41.2 bits (95), Expect = 0.005
Identities = 30/124 (24%), Positives = 50/124 (40%)
Frame = +3
Query: 1463 DNTGIEMDQDFQADAVSLSEDSSENEDCDGENEELESEMGPTGPDSEAVGEKIWDQNEDE 1522
D G E+ Q D + ++SS ++DC +N + + ++ D +G ++DE
Sbjct: 723 DVPGAELLQIIDDD---VEQESSHSDDCGSDNAQEDDQVSLNSDDDNQLGSDNTGSDDDE 893
Query: 1523 TPDDTGEKYESGPSVKDGDGSNKELRAKDDSTSDQSGDDSCDEGDAQNDEAAAQNEFDDE 1582
D DG ++ R+ D TS D+ DEGD D + E
Sbjct: 894 AED------------HDGVSDDENDRSSDYETSGDDADNVEDEGDDLEDSEEDGGISEHE 1037
Query: 1583 ENGD 1586
+GD
Sbjct: 1038 GDGD 1049
Score = 36.2 bits (82), Expect = 0.15
Identities = 34/120 (28%), Positives = 50/120 (41%), Gaps = 7/120 (5%)
Frame = +3
Query: 1443 LLSMMQKKEEQEES-KEVPGNDNT------GIEMDQDFQADAVSLSEDSSENEDCDGENE 1495
LL ++ EQE S + G+DN + D D Q + + D E ED DG ++
Sbjct: 741 LLQIIDDDVEQESSHSDDCGSDNAQEDDQVSLNSDDDNQLGSDNTGSDDDEAEDHDGVSD 920
Query: 1496 ELESEMGPTGPDSEAVGEKIWDQNEDETPDDTGEKYESGPSVKDGDGSNKELRAKDDSTS 1555
+ D E G+ D EDE D + + G S +GDG L + D T+
Sbjct: 921 DENDRSS----DYETSGDDA-DNVEDEGDDLEDSEEDGGISEHEGDGDLHILGSVDTKTT 1085
Score = 34.7 bits (78), Expect = 0.44
Identities = 32/135 (23%), Positives = 59/135 (43%), Gaps = 11/135 (8%)
Frame = +3
Query: 1643 QEDETCPPD-EIMEEAHTEVDVSSE----------KDDLGQEHQENDDMNSMDPKNDTSE 1691
++D P D + +A+ EV+V+++ DD+ QE +DD S D ++
Sbjct: 651 KKDRGRPTDPKAKPKAYGEVNVAADVPGAELLQIIDDDVEQESSHSDDCGS-----DNAQ 815
Query: 1692 SSDVVNPQVSNVDLASQSKSDLQTSGSENIASQSNLSNSHHDFGNPAVTGGFPSSDMSEM 1751
D V+ N D +Q SD S + +S+ +D + T G + ++ +
Sbjct: 816 EDDQVS---LNSDDDNQLGSDNTGSDDDEAEDHDGVSDDENDRSSDYETSGDDADNVEDE 986
Query: 1752 DINMSDSSNTGGFSK 1766
++ DS GG S+
Sbjct: 987 GDDLEDSEEDGGISE 1031
Score = 34.3 bits (77), Expect = 0.58
Identities = 30/97 (30%), Positives = 43/97 (43%), Gaps = 3/97 (3%)
Frame = +3
Query: 1541 DGSNKELRAKDDSTSDQSGDDS---CDEGDAQNDEAAAQNEFDDEENGDDVNMDKEAAHS 1597
D + +DD S S DD+ D + +DEA + D+EN D + D E +
Sbjct: 789 DDCGSDNAQEDDQVSLNSDDDNQLGSDNTGSDDDEAEDHDGVSDDEN--DRSSDYETSGD 962
Query: 1598 DATGLKPDEPDHSSDMEIDLNANEDVDPIEEGDQEGH 1634
DA ++ DE D D E D +E EGD + H
Sbjct: 963 DADNVE-DEGDDLEDSEEDGGISE-----HEGDGDLH 1055
>TC79717 similar to GP|10086499|gb|AAG12559.1 Unknown Protein {Arabidopsis
thaliana}, partial (5%)
Length = 1579
Score = 41.2 bits (95), Expect = 0.005
Identities = 83/375 (22%), Positives = 131/375 (34%), Gaps = 61/375 (16%)
Frame = +2
Query: 1493 ENEELESEMG----PTGPDSEAVGEKIW--------DQNEDETPDDT-GEKYESGPSVKD 1539
+NE +ES P P ++ +GE+ DQ E E T G + S PS
Sbjct: 188 QNEIMESNASVGHEPNSPQNQEIGEQYKLSTKAFASDQLETEGKKITFGSRKLSNPSFIA 367
Query: 1540 GDGSNKELRAKDDSTSDQSGDDSCDEGDAQNDEAAAQNEFDDEENGDDV---NMDKEAAH 1596
+EL + +S D ++Q D A EF EN N D E+
Sbjct: 368 AQSKFEELSSNSNSGRPSGLLDQDVSVESQADSAYISKEFISSENSTPYPSRNADPESGT 547
Query: 1597 SDATGLKPDEPDHSSDMEIDLNANEDVD----------------------PIEEGDQEGH 1634
+ D PD S +EI+ +A + V+ PI + DQ
Sbjct: 548 VLSISSTLDSPDRSETLEIEHDAKDLVEGIVNPENKTDHGVEANTPTSNLPISDSDQL-E 724
Query: 1635 DDSAENGNQEDETCPP---DEIMEEAHTEVDVSSEK-----DDLGQEHQENDDMNSMDPK 1686
+ GN D P + +E T D+ E+ D Q + P+
Sbjct: 725 TVNGSRGNVVDSVMPENSKEHAVEPEKTASDLLREQTETVLQDFTYSQQASPGSYMTIPE 904
Query: 1687 NDTSESSDV--------VNP-------QVSNVDLASQSKSDLQTSGSENIASQSNLSNSH 1731
+ + SS V +N +V +V S + S+ Q SGS + +N++
Sbjct: 905 SQGTPSSQVSVKTKENKINKTGSSSRRRVLSVGNKSPANSN-QDSGSRVFVGTKSPANAN 1081
Query: 1732 HDFGNPAVTGGFPSSDMSEMDINMSDSSNTGGFSKTQPKSHLPQHEHSFSQEKQTNPSRS 1791
HD G+ P ++ N+ G K + P ++S S ++
Sbjct: 1082 HDSGSRGSREQLPKDQLN------GKRRNSFGSIKPEHTDQEPTKDNSSSNNTLPRFMQA 1243
Query: 1792 TGDALDFRKEKINVS 1806
T A K KIN +
Sbjct: 1244 TQSA----KAKINAN 1276
>TC87847 similar to GP|15929245|gb|AAH15068.1 Similar to adaptor-related
protein complex AP-3 beta 1 subunit {Mus musculus},
partial (2%)
Length = 931
Score = 41.2 bits (95), Expect = 0.005
Identities = 51/223 (22%), Positives = 87/223 (38%), Gaps = 29/223 (13%)
Frame = +1
Query: 1610 SSDMEIDLNANEDVDPIEEGDQEGHDDSAENGNQEDETCPPDEIMEEAHTEVDVSSE--- 1666
SSD E + + D + E E G++ED + DE E+ T+ S +
Sbjct: 115 SSDEEQPPSTQKPADKVVNEQDEDSSSEEEEGDEEDGSSSEDEEEEDNQTQQTPSKDSKP 294
Query: 1667 -----------------KDDLGQEHQENDDMNSMDPKNDTSESSDVVNPQV---SNVDLA 1706
+ + G E S SE + NP+V ++ +
Sbjct: 295 ASKNPPPSTPISNPKPAESESGSESGSESGTESDSEPEHKSEPTPPPNPKVKPLASKPMK 474
Query: 1707 SQSKSDLQT------SGSENIASQSNLSNSHHDFGNPAVTGGFPSSDMSEMDINMSDSSN 1760
+Q+++ Q+ SG++ +A +S+ N T G S D +E++ D+
Sbjct: 475 TQTQAQAQSTPVPIKSGTKRVA-ESSTGNDSKRSKKKTTTAGGGSDDENEVE---EDAKL 642
Query: 1761 TGGFSKTQPKSHLPQHEHSFSQEKQTNPSRSTGDALDFRKEKI 1803
TG SK + FS+E++T S+ T L R+EKI
Sbjct: 643 TGEDSK-------KNFQRVFSEEERTCDSQRTWGILWRRQEKI 750
>BG644339 weakly similar to PIR|T48401|T484 histone deacetylase-like protein -
Arabidopsis thaliana, partial (26%)
Length = 763
Score = 40.8 bits (94), Expect = 0.006
Identities = 37/133 (27%), Positives = 56/133 (41%), Gaps = 7/133 (5%)
Frame = +1
Query: 1434 EHVNRYVNCLLSMMQKKEEQEESKEVPGNDNTGIEMDQDFQADAVSLSEDSSENEDCDGE 1493
E+V +N L K + + K P D ++ A S E S+ +D D E
Sbjct: 361 ENVLEALNGKLEADVKDVKPDAKKSAPVKDEKNAKI-------AASKKEAESDEDDSDSE 519
Query: 1494 NEELESEMGPTGP-------DSEAVGEKIWDQNEDETPDDTGEKYESGPSVKDGDGSNKE 1546
+ + +G GP DSE + D +E+ETP +K PS K GS K
Sbjct: 520 GDS-DVPLGMDGPEGMDFSGDSEDDDDSEEDDSEEETPKKVEQKKRPAPSPKVAPGSGK- 693
Query: 1547 LRAKDDSTSDQSG 1559
+AK + + +SG
Sbjct: 694 -KAKQVTPAHKSG 729
>BE203389 weakly similar to PIR|T06754|T0 DNA-directed RNA polymerase I 190K
chain homolog F15B8.150 - Arabidopsis thaliana, partial
(6%)
Length = 652
Score = 40.4 bits (93), Expect = 0.008
Identities = 26/102 (25%), Positives = 47/102 (45%), Gaps = 1/102 (0%)
Frame = +2
Query: 1545 KELRAKDDSTSDQSGDDSCDEGDAQNDEAAAQNEFDDEENGDDVNMDKEAAHSDATGLKP 1604
K+ + K +S++ D S + QN + +E D E+ ++ D + + AT +
Sbjct: 329 KDFQGKSNSSNGLDNDHSNESASNQNGQTDDDDEVGDTEDAEEDGFDAQKSKQRATD-EV 505
Query: 1605 DEPDHSSDMEIDLNANEDVDPIEEGDQEGHDDSAE-NGNQED 1645
D D + D +EDV+ E+G + D+ E NG+ D
Sbjct: 506 DYDDGPEEETHDGEKSEDVEVSEDGKDDEDDNGVEVNGDDSD 631
Score = 35.8 bits (81), Expect = 0.20
Identities = 30/131 (22%), Positives = 53/131 (39%)
Frame = +2
Query: 1457 KEVPGNDNTGIEMDQDFQADAVSLSEDSSENEDCDGENEELESEMGPTGPDSEAVGEKIW 1516
K+ G N+ +D D ++ S ++++D G+ E+ E + G D++ ++
Sbjct: 329 KDFQGKSNSSNGLDNDHSNESASNQNGQTDDDDEVGDTEDAEED----GFDAQKSKQRAT 496
Query: 1517 DQNEDETPDDTGEKYESGPSVKDGDGSNKELRAKDDSTSDQSGDDSCDEGDAQNDEAAAQ 1576
D+ + Y+ GP + DG E D S+ DD D G N +
Sbjct: 497 DEVD----------YDDGPEEETHDGEKSE----DVEVSEDGKDDEDDNGVEVNGD---- 622
Query: 1577 NEFDDEENGDD 1587
+ D E N D
Sbjct: 623 -DSDIEVNDSD 652
>TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {Arabidopsis
thaliana}, partial (39%)
Length = 883
Score = 39.7 bits (91), Expect = 0.014
Identities = 23/80 (28%), Positives = 36/80 (44%), Gaps = 10/80 (12%)
Frame = +3
Query: 1580 DDEENGDDVNMDKEAAHSDATGLKPDEPDHSSDMEIDLNAN----------EDVDPIEEG 1629
DDE++ DD ++ E + DE + D E D AN +D D EE
Sbjct: 327 DDEDDDDDDDVQDEDDDGEEEDYSGDEGEEEGDPEDDPEANGAGGSDDGEDDDDDGDEED 506
Query: 1630 DQEGHDDSAENGNQEDETCP 1649
+++G D+ E +ED+ P
Sbjct: 507 EEDGEDEEDEEDEEEDDETP 566
Score = 38.9 bits (89), Expect = 0.024
Identities = 31/112 (27%), Positives = 49/112 (43%), Gaps = 3/112 (2%)
Frame = +3
Query: 1506 PDSEAVGEK-IWDQNEDETPDDTGEKYESGPSVKDGDGSNKELRAKDDSTSDQSGDDSCD 1564
P +G K + +N+ +T DD + + +D DG + D SGD+ +
Sbjct: 267 PLEHLLGRKPAYKENKSDTEDDEDDDDDDDVQDEDDDGEEE----------DYSGDEGEE 416
Query: 1565 EGDAQNDEAA--AQNEFDDEENGDDVNMDKEAAHSDATGLKPDEPDHSSDME 1614
EGD ++D A A D E++ DD + + E D + DE D D E
Sbjct: 417 EGDPEDDPEANGAGGSDDGEDDDDDGDEEDEEDGED----EEDEEDEEEDDE 560
Score = 35.0 bits (79), Expect = 0.34
Identities = 21/85 (24%), Positives = 38/85 (44%), Gaps = 2/85 (2%)
Frame = +3
Query: 1517 DQNEDETPDDTGEKYESGPSVKDGDGSNKELRAKDDSTSDQSGDDSCDEGDAQNDEAAAQ 1576
D +D+ DD ++ + G +D G E + + +G D+G+ +D+ +
Sbjct: 327 DDEDDDDDDDVQDEDDDGEE-EDYSGDEGEEEGDPEDDPEANGAGGSDDGEDDDDDGDEE 503
Query: 1577 NEFD--DEENGDDVNMDKEAAHSDA 1599
+E D DEE+ +D D E A
Sbjct: 504 DEEDGEDEEDEEDEEEDDETPQPPA 578
Score = 32.0 bits (71), Expect = 2.9
Identities = 31/115 (26%), Positives = 47/115 (39%), Gaps = 2/115 (1%)
Frame = +3
Query: 1573 AAAQNEFDDEENGDDVNMDKEAAHSDATGLKPDEPDHSSDMEIDLNANEDVDPIEEGDQE 1632
A +N+ D E++ DD + D + D G +E D+S D EG++E
Sbjct: 297 AYKENKSDTEDDEDDDD-DDDVQDEDDDG---EEEDYSGD---------------EGEEE 419
Query: 1633 G--HDDSAENGNQEDETCPPDEIMEEAHTEVDVSSEKDDLGQEHQENDDMNSMDP 1685
G DD NG + D+ + E D E+D +E +E DD P
Sbjct: 420 GDPEDDPEANGAGGSDDGEDDDDDGDEEDEEDGEDEED---EEDEEEDDETPQPP 575
>TC82499 SP|Q9XHM1|IF38_MEDTR Eukaryotic translation initiation factor 3
subunit 8 (eIF3 p110) (eIF3c). [Barrel medic], partial
(91%)
Length = 2816
Score = 39.3 bits (90), Expect = 0.018
Identities = 40/168 (23%), Positives = 71/168 (41%), Gaps = 13/168 (7%)
Frame = +1
Query: 1424 IR*LMKISCWEHVNRYVNCLLSMMQKKEEQEESKEVPGNDNTGIEMDQDFQADAVSLSED 1483
I+ +KI+ W + + + ++K ES+++P + M +DF A A S ++D
Sbjct: 475 IKNAIKINDWVSLQESFDKINKQLEKVMRVIESQKIPNLYIKALVMLEDFLAQA-SANKD 651
Query: 1484 S-------------SENEDCDGENEELESEMGPTGPDSEAVGEKIWDQNEDETPDDTGEK 1530
+ S + N++ E + E+ GEK DE +D+ E
Sbjct: 652 AKKKMSPSNAKAFNSMKQKLKKNNKQYEDLIIKCRESPESEGEK------DEDDEDSDE- 810
Query: 1531 YESGPSVKDGDGSNKELRAKDDSTSDQSGDDSCDEGDAQNDEAAAQNE 1578
YES + + D K D TS+ D D+GDA D+ ++ +
Sbjct: 811 YESDDEMIEPDQLRKPEPVSDSETSELGNDRPGDDGDAPWDQKLSKKD 954
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum}, partial
(13%)
Length = 663
Score = 39.3 bits (90), Expect = 0.018
Identities = 38/144 (26%), Positives = 61/144 (41%), Gaps = 11/144 (7%)
Frame = +3
Query: 1514 KIWDQNEDETPDDTGE---KYESGPSVKDGDGSNKELRAKDDSTSDQSGDDSCDEGDAQN 1570
KI D++ + +D E E KD + K+ + K D T G D D+ + +
Sbjct: 90 KIDDKSAGDVKEDKVEIEKDLEIKSVEKDDEKKEKKDKEKKDKTDVDEGKDKKDK-EKKK 266
Query: 1571 DEAAAQNEFDDEENGDDVNMDKEAAHSDATGLKPDEPDHSS-------DMEIDLNANEDV 1623
E +N +EE+GD+ DKE + ++ D D E + NED
Sbjct: 267 KEKKEENVKGEEEDGDE-KKDKEKKKKEKKEKGKEDKDKDGEEKKSKKDKEKKKDKNEDD 443
Query: 1624 DPIEEGD-QEGHDDSAENGNQEDE 1646
D E+G ++ + D E +EDE
Sbjct: 444 DEGEDGSKKKKNKDKKEKKKEEDE 515
Score = 37.7 bits (86), Expect = 0.053
Identities = 32/146 (21%), Positives = 60/146 (40%)
Frame = +3
Query: 1448 QKKEEQEESKEVPGNDNTGIEMDQDFQADAVSLSEDSSENEDCDGENEELESEMGPTGPD 1507
+KK+++++ + V G + G E +D + E E++D DGE ++ + +
Sbjct: 255 EKKKKEKKEENVKGEEEDGDEK-KDKEKKKKEKKEKGKEDKDKDGEEKKSKKDK------ 413
Query: 1508 SEAVGEKIWDQNEDETPDDTGEKYESGPSVKDGDGSNKELRAKDDSTSDQSGDDSCDEGD 1567
EK D+NED+ + G K + K+ E S D +++ EG
Sbjct: 414 -----EKKKDKNEDDDEGEDGSKKKKNKDKKEKKKEEDEKEEGKVSVRDIDIEETAKEGK 578
Query: 1568 AQNDEAAAQNEFDDEENGDDVNMDKE 1593
+ + + D EE DK+
Sbjct: 579 EKK-----KKKEDKEEKKKKSGKDKD 641
Score = 35.4 bits (80), Expect = 0.26
Identities = 27/148 (18%), Positives = 61/148 (40%), Gaps = 6/148 (4%)
Frame = +3
Query: 1571 DEAAAQNEFDDEENGD------DVNMDKEAAHSDATGLKPDEPDHSSDMEIDLNANEDVD 1624
+E+ + DD+ GD ++ D E + K ++ D + D++ +D
Sbjct: 69 EESNIPIKIDDKSAGDVKEDKVEIEKDLEIKSVEKDDEKKEKKDKEKKDKTDVDEGKDKK 248
Query: 1625 PIEEGDQEGHDDSAENGNQEDETCPPDEIMEEAHTEVDVSSEKDDLGQEHQENDDMNSMD 1684
E+ +E +++ + G +ED D+ ++ + +KD G+E + D
Sbjct: 249 DKEKKKKEKKEENVK-GEEEDGDEKKDKEKKKKEKKEKGKEDKDKDGEEKKSKKDKEKKK 425
Query: 1685 PKNDTSESSDVVNPQVSNVDLASQSKSD 1712
KN+ + + + + N D + K +
Sbjct: 426 DKNEDDDEGEDGSKKKKNKDKKEKKKEE 509
Score = 33.5 bits (75), Expect = 0.99
Identities = 38/176 (21%), Positives = 69/176 (38%), Gaps = 8/176 (4%)
Frame = +3
Query: 1543 SNKELRAKDDSTSDQSGDDSCDEGDAQNDEAAAQNEFDDEENGDDVNMDKEAAHSDATGL 1602
SN ++ D S +GD D+ + + D E DDE+ K+ D
Sbjct: 75 SNIPIKIDDKS----AGDVKEDKVEIEKDLEIKSVEKDDEKKEKKDKEKKDKTDVDEGKD 242
Query: 1603 KPDEPDHSSDMEIDLNANEDVDPIEEGDQEGHDDSAENGNQEDETCPPDEIMEEAHTEVD 1662
K D+ + + + E+ D E+ D+E + +ED+ D+ EE ++ D
Sbjct: 243 KKDKEKKKKEKKEENVKGEEEDGDEKKDKEKKKKEKKEKGKEDK----DKDGEEKKSKKD 410
Query: 1663 VSSEKD--------DLGQEHQENDDMNSMDPKNDTSESSDVVNPQVSNVDLASQSK 1710
+KD + G + ++N D + D E V V ++D+ +K
Sbjct: 411 KEKKKDKNEDDDEGEDGSKKKKNKDKKEKKKEEDEKEEGKV---SVRDIDIEETAK 569
>TC85559 similar to PIR|T07012|T07012 acetyl-CoA carboxylase (EC 6.4.1.2) -
potato chloroplast, partial (66%)
Length = 5011
Score = 39.3 bits (90), Expect = 0.018
Identities = 25/116 (21%), Positives = 53/116 (45%), Gaps = 1/116 (0%)
Frame = +3
Query: 1519 NEDETPDDTGEKYESGPSVKDGDGSNKELRAKD-DSTSDQSGDDSCDEGDAQNDEAAAQN 1577
N ++ P + G + E PS + E +D D S+ S +D E ++++ ++
Sbjct: 3462 NSEDEPSEKGLEDEDEPSENSLEDEPSENSLEDEDEPSENSLEDEPSEKGLEDEDEPSEK 3641
Query: 1578 EFDDEENGDDVNMDKEAAHSDATGLKPDEPDHSSDMEIDLNANEDVDPIEEGDQEG 1633
+DE+ + ++ E S+ + DEP + D + L++ +D + + Q G
Sbjct: 3642 GLEDEDEPSEKGLEDEDEPSENSLEDEDEPSENDDYQNRLDSYQDRTGLLDAVQTG 3809
Score = 30.8 bits (68), Expect = 6.4
Identities = 13/57 (22%), Positives = 29/57 (50%)
Frame = +3
Query: 1635 DDSAENGNQEDETCPPDEIMEEAHTEVDVSSEKDDLGQEHQENDDMNSMDPKNDTSE 1691
D ++GN +D+ +I+E + + S+ D+ ++ +DD +++ DT E
Sbjct: 2712 DKEKDSGNDDDKLQKASDILEPVNYSAENYSDSDNDSDDNDPDDDYDTLQKGTDTLE 2882
>BQ140761 weakly similar to GP|18076245|emb phosphophoryn {Rattus norvegicus},
partial (9%)
Length = 620
Score = 39.3 bits (90), Expect = 0.018
Identities = 43/189 (22%), Positives = 74/189 (38%), Gaps = 18/189 (9%)
Frame = +1
Query: 1477 AVSLSEDSSENEDCDGENEELESEMGPTGPDSEA---VGEKIWD-----QNEDETPDDTG 1528
A S + ++ ED + + ++S + TG D A G W Q++ D+
Sbjct: 67 AKSDKKSATAPEDVQTDLDLVQSFLKQTGHDKAASTLTGFAAWATKQAAQSDAMDVDEKS 246
Query: 1529 EKYESGPSVKDGDGSNKELRAKDDS--TSDQSGDDSCDEGD---AQNDEAA-----AQNE 1578
S DGS+ + A DDS +SD S DDS + + A E A A++
Sbjct: 247 SDSSDSDSSDSSDGSDSDSDASDDSDDSSDSSSDDSSSDEEPVPAPKKEKAKKAKKAKSV 426
Query: 1579 FDDEENGDDVNMDKEAAHSDATGLKPDEPDHSSDMEIDLNANEDVDPIEEGDQEGHDDSA 1638
++ DD + + E+ P+ D SS A+ D D + + DS+
Sbjct: 427 SSSSDSSDDSSSESESEPEPXNVPLPESDDSSS-------ASSDSDSSSDSSSDSSSDSS 585
Query: 1639 ENGNQEDET 1647
+ + +
Sbjct: 586 SXSDXDSSS 612
>TC93063 similar to PIR|T08852|T08852 lustrin A - California red abalone,
partial (4%)
Length = 1058
Score = 38.9 bits (89), Expect = 0.024
Identities = 44/222 (19%), Positives = 90/222 (39%), Gaps = 6/222 (2%)
Frame = -1
Query: 1561 DSCDEGDAQNDEAAAQNEFDDEENGDDVNMDKEAAHSDATGLKPDEPDHSSDMEIDLNAN 1620
D C+EGD Q + + + N V++ +DA EP+ + E +
Sbjct: 749 DMCEEGDLQCQYPLVKKQAPRKSN---VSIGAAEDEADAEADDEPEPEPEPEPEAEPEPE 579
Query: 1621 EDVDPIEEGDQEGHDDSAENGNQEDETCPPDEIMEEAHTEVDVSSEKDDLGQEHQENDDM 1680
+ +P E + E + E E P DE E+ +++ ++ + +
Sbjct: 578 AEPEPEAEPEPEAEPE------PEAEPEPEDEEEEQEPDDIETIDYTSNMPVSNMLTPAV 417
Query: 1681 NSMDPKNDTSESSDVVNPQVSNVDLA-SQSKSDLQTSGSENIASQSNLSNSHHDFGNPAV 1739
+ + S SD+ PQ S D+A S S + + + + Q ++ S +G PA+
Sbjct: 416 ETANQDYFNSAPSDLTYPQPSAHDIAMSAVGSYPDPAATVDYSDQHAVTTS---YGQPAL 246
Query: 1740 T-----GGFPSSDMSEMDINMSDSSNTGGFSKTQPKSHLPQH 1776
T G +++ S+ + ++ ++ G + P++ QH
Sbjct: 245 TETGSYQGAVNTNYSQQAVMDTNGASLGQSATKSPRAARAQH 120
>BQ149637 similar to PIR|D85135|D85 hypothetical protein AT4g12610 [imported] -
Arabidopsis thaliana, partial (15%)
Length = 538
Score = 33.5 bits (75), Expect = 0.99
Identities = 34/162 (20%), Positives = 65/162 (39%), Gaps = 2/162 (1%)
Frame = +3
Query: 1448 QKKEEQEESKEVPGNDNTGIEMDQDFQADAVSLSEDSSENEDCDGENEELESEMGPTGPD 1507
+ ++E+ + K+ P NT + D++ +D + D E+EE+ ++ D
Sbjct: 24 EDEDEEVDRKKRPNKKNT-FDDDEEGPRGGDHDEDDYDVEKGDDWEHEEIFTD------D 182
Query: 1508 SEAVGEKIWDQNEDETPDDTGEKYESGPSVKDGDGSNKELRAKDDSTSDQSGDDSCDEGD 1567
EAVG N+ E +D + + P +K D +E D +++ G S +
Sbjct: 183 DEAVG------NDPEEREDLAPEVPAPPEIKQDDEEEEE-----DEXNEEGGGLSKSGKE 329
Query: 1568 AQN--DEAAAQNEFDDEENGDDVNMDKEAAHSDATGLKPDEP 1607
+ +E DD+++ D +D T EP
Sbjct: 330 LKKLLGRTGGLSESDDDDDXXDDXIDDXXGVPAVTATXXXEP 455
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.133 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 69,491,830
Number of Sequences: 36976
Number of extensions: 972038
Number of successful extensions: 5636
Number of sequences better than 10.0: 89
Number of HSP's better than 10.0 without gapping: 5342
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5596
length of query: 2332
length of database: 9,014,727
effective HSP length: 112
effective length of query: 2220
effective length of database: 4,873,415
effective search space: 10818981300
effective search space used: 10818981300
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC148398.1


