

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148396.11 + phase: 0
(745 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
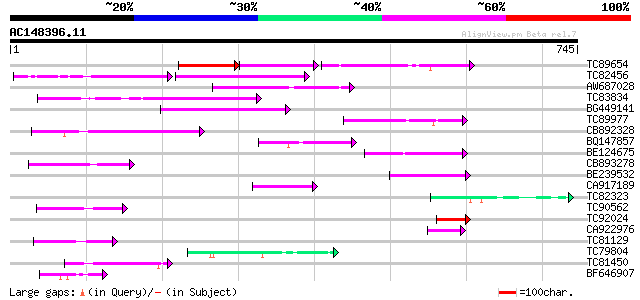
Score E
Sequences producing significant alignments: (bits) Value
TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein... 112 2e-65
TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far... 145 2e-42
AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~... 146 2e-35
TC83834 similar to PIR|G96565|G96565 F6D8.26 [imported] - Arabid... 120 2e-27
BG449141 similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidop... 105 8e-23
TC89977 similar to GP|5764395|gb|AAD51282.1| far-red impaired re... 100 3e-21
CB892328 GP|15075873|em PROBABLE GLYCOSYL HYDROLASE PROTEIN {Sin... 93 3e-19
BQ147857 weakly similar to GP|15983442|gb At1g10240/F14N23_12 {A... 76 5e-14
BE124675 weakly similar to GP|13365573|db hypothetical protein~s... 70 3e-12
CB893278 weakly similar to PIR|T05645|T056 hypothetical protein ... 68 1e-11
BE239532 64 3e-10
CA917189 similar to PIR|T05645|T05 hypothetical protein F20D10.3... 63 4e-10
TC82323 similar to GP|7673677|gb|AAF66982.1| transposase {Zea ma... 57 2e-08
TC90562 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20... 57 3e-08
TC92024 similar to PIR|T05645|T05645 hypothetical protein F20D10... 55 8e-08
CA922976 weakly similar to GP|9502168|gb|A contains simlarity to... 49 9e-06
TC81129 similar to GP|5764395|gb|AAD51282.1| far-red impaired re... 45 8e-05
TC79804 weakly similar to GP|6175165|gb|AAF04891.1| Mutator-like... 42 7e-04
TC81450 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20... 42 9e-04
BF646907 similar to GP|15983442|gb| At1g10240/F14N23_12 {Arabido... 42 9e-04
>TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein F20D10.290
- Arabidopsis thaliana, partial (65%)
Length = 1358
Score = 112 bits (280), Expect(3) = 2e-65
Identities = 71/207 (34%), Positives = 103/207 (49%), Gaps = 6/207 (2%)
Frame = +1
Query: 410 NGWLSTMYDLRSMWIPAYFKDTFMAGILRTTSRSESENSFFGNYLNHNLTLVEFWVRFDS 469
N W +MY+ R W+P Y + +F G + +E N FF ++N + TL +++
Sbjct: 697 NEWPQSMYNARQHWVPVYLRGSFF-GEIPLNDGNECLNFFFDGHVNASTTLQLLVRQYEK 873
Query: 470 ALAAQRHKELFADNNTLHSNPELNMHMNLEKHAREVYTHENFYIFQKELWSACVDCGIEG 529
A++ +EL AD T +SNP L +EK A +YT + F FQ+EL +
Sbjct: 874 AVSTWHERELKADFETSNSNPVLRTPSPMEKQAASLYTRKMFMKFQEELVGTMAN---PA 1044
Query: 530 TKEEGENLSFSILDNAVRKHRE------VVYCLSNNIAHCSCKMFESEGIPCRHILFILK 583
TK E S +I V K E V + S A CSC+MFE GI CRHIL + +
Sbjct: 1045TKIED---SGTITTYRVAKFGENQTSHTVAFNSSEMKASCSCQMFEYSGIVCRHILAVFR 1215
Query: 584 GKGFSEIPSHYIVNRWTKLATSKPIFD 610
K +PSHY++ RWT A + + D
Sbjct: 1216AKNVLTLPSHYVLKRWTMNAKTGVVLD 1296
Score = 91.3 bits (225), Expect(3) = 2e-65
Identities = 41/80 (51%), Positives = 53/80 (66%)
Frame = +2
Query: 223 IDNFRRKREINPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYRTNKYFM 282
+D +R R N +FFY ++D + ++FWAD CR NYS FGD V FDTTY+TN+Y +
Sbjct: 134 LDYLKRMRAENHAFFYAVQSDVDNAGGNIFWADETCRTNYSYFGDTVIFDTTYKTNQYRV 313
Query: 283 IFAPFTGINHHRQSITFGAA 302
FA FTG NHH Q + FG A
Sbjct: 314 PFASFTGFNHHGQPVLFGCA 373
Score = 86.3 bits (212), Expect(3) = 2e-65
Identities = 39/103 (37%), Positives = 61/103 (58%)
Frame = +3
Query: 303 LLKNEKEESFVWLFETFLKAMGGHKPVMIITDQDGGMKNAIGAVLKGSSHRFCMWHILKK 362
L+ NE E S++WLF+T+L+A+ G PV I TD D ++ A+ VL + HRFC W I ++
Sbjct: 375 LILNESEPSYIWLFKTWLRAVSGRPPVSITTDLDPVIQVAVAQVLPPTRHRFCKWSIFRE 554
Query: 363 LSEKVGSSMDENSGFNDRFKSCVWNSESSEEFDLEWNNIISDY 405
K+ N F++ FK CV S + +EF+ W +++ Y
Sbjct: 555 NRSKLAHLYQSNPTFDNEFKKCVHESVTIDEFESCWRSLLERY 683
>TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far-red
impaired response protein {Oryza sativa}, partial (7%)
Length = 1244
Score = 145 bits (367), Expect(2) = 2e-42
Identities = 68/177 (38%), Positives = 102/177 (57%)
Frame = +2
Query: 218 DADMFIDNFRRKREINPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYRT 277
D D F R ++ N F+ + D++ ++++FWAD CR Y FG+V++ DTTY T
Sbjct: 716 DVDAIHSFFHRMQKQNSQFYCAMDMDDKRNIQNLFWADARCRAAYEYFGEVITLDTTYLT 895
Query: 278 NKYFMIFAPFTGINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGHKPVMIITDQDG 337
+KY + PF G+NHH Q+I G A+L N ++ WLF +L+ M GH P IIT++D
Sbjct: 896 SKYDLPLVPFVGVNHHGQTILLGCAILSNLDAKTLTWLFTRWLECMHGHAPNGIITEEDK 1075
Query: 338 GMKNAIGAVLKGSSHRFCMWHILKKLSEKVGSSMDENSGFNDRFKSCVWNSESSEEF 394
MKNAI + HR+C+WHI+KK+ E +G N + V++S S +F
Sbjct: 1076AMKNAIEVAFPKARHRWCLWHIMKKVPEMLG-KYSRNESIKTLLQDVVYDSMSKSDF 1243
Score = 45.8 bits (107), Expect(2) = 2e-42
Identities = 48/208 (23%), Positives = 89/208 (42%)
Frame = +1
Query: 6 NEVDFSSPSKDGNDPKNTSVEWIPACEDELKPVIGKVFDTLVEGGDFYKAYAYVAGFSVR 65
++V+ S SK +D + + I EDE +P G F + E +Y YA GF V
Sbjct: 127 DKVEKSEASKRSDDKFSAA---IVVSEDE-EPRTGMTFRSEEEVIRYYMNYANRVGFGV- 291
Query: 66 NSIKTKDKDGVKWKYFLCSKEGFKEEKKVDKPQLLIAENSLSKSRKRKLTREGCKARLVL 125
I +K+ D K KYF + + K + S++ ++ C+ARL
Sbjct: 292 TKISSKNADDGK-KYFTLACNCARRYVSTSK----------NPSKQYLTSKTQCRARLNA 438
Query: 126 KRTIDGKYEVSNFYEGHSHGLVSPSKRQFLRSARNVTSVHKNILFSCNRANVGTSKSYQI 185
+DG +S H+H L R F + T V +N+ + ++ + ++ Q
Sbjct: 439 CVALDGSSTISRIVLEHNHELGQTKGRYFRLDESSGTHVERNLELN-DQGGINVDRNLQS 615
Query: 186 MKEQVGSYENIGCTQRDLQNYSRNLKEL 213
+ ++ N+ + D +N+ + ++ L
Sbjct: 616 VGLEMNGCGNLTSGENDCRNFVQKVRRL 699
>AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (11%)
Length = 662
Score = 146 bits (369), Expect = 2e-35
Identities = 72/187 (38%), Positives = 109/187 (57%)
Frame = +1
Query: 267 DVVSFDTTYRTNKYFMIFAPFTGINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGH 326
DV++FDTTY+ NKY F+G NHH Q+I FG ALL++E ES+ W+ FL+ M
Sbjct: 1 DVLAFDTTYKKNKYNYPLCIFSGCNHHSQTIIFGVALLEDETIESYKWVLNRFLECMENK 180
Query: 327 KPVMIITDQDGGMKNAIGAVLKGSSHRFCMWHILKKLSEKVGSSMDENSGFNDRFKSCVW 386
P ++TD DG M+ AI V +SHR C WH+ K E + + + F + F+ ++
Sbjct: 181 FPKAVVTDGDGSMREAIKQVFPDASHRLCAWHLHKNAQENI-----KKTPFLEGFRKAMY 345
Query: 387 NSESSEEFDLEWNNIISDYSLEGNGWLSTMYDLRSMWIPAYFKDTFMAGILRTTSRSESE 446
++ + E+F+ W+ +I LEGN W+ Y +S+W AY +D F G +RTTS+ E+
Sbjct: 346 SNFTPEQFEDFWSELIQKNELEGNAWVIKTYANKSLWATAYLRDKFF-GRIRTTSQCEAI 522
Query: 447 NSFFGNY 453
N+ Y
Sbjct: 523 NAIVKTY 543
>TC83834 similar to PIR|G96565|G96565 F6D8.26 [imported] - Arabidopsis
thaliana, partial (24%)
Length = 1032
Score = 120 bits (300), Expect = 2e-27
Identities = 89/298 (29%), Positives = 131/298 (43%), Gaps = 3/298 (1%)
Frame = +3
Query: 37 PVIGKVFDTLVEGGDFYKAYAYVAGFSVR--NSIKTKDKDGVKWKYFLCSKEGFKEEKKV 94
P + F++ + +Y YA GF VR NS ++ CS +GFK K V
Sbjct: 213 PALAMEFESYDDAYSYYICYAKEVGFCVRVKNSWFKRNSKEKYGAVLCCSSQGFKRTKDV 392
Query: 95 DKPQLLIAENSLSKSRKRKLTREGCKARLVLKRTIDGKYEVSNFYEGHSHGLVSPSKRQF 154
N+L RK TR GC A + +K ++ + H+H L
Sbjct: 393 ---------NNL-----RKETRTGCPAMIRMKLVESQRWRICEVTLEHNHVL-------- 506
Query: 155 LRSARNVTSVHKNILFSCNRANVGTSKSYQIMKEQVGSYENIGCTQRDLQNYSRNLKEL- 213
A+ S+ KN L S + A T K Y + +N+ RD + +S+ +L
Sbjct: 507 --GAKIHKSIKKNSLPSSD-AEGKTIKVYHALVIDTEGNDNLNSNARDDRAFSKYSNKLN 677
Query: 214 IKDSDADMFIDNFRRKREINPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDT 273
++ D + R + NP+FFY + ++EG L++ W D R F DV+ FD
Sbjct: 678 LRKGDTQAIYNFLCRMQLTNPNFFYLMDFNDEGHLRNALWVDAKSRAACGYFSDVIYFDN 857
Query: 274 TYRTNKYFMIFAPFTGINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGHKPVMI 331
TY NKY + GINHH QS+ G LL E ES+ WLF T++K + P I
Sbjct: 858 TYLVNKYEIPLVALVGINHHGQSVLLGCGLLAGEIIESYKWLFRTWIKCIPRCSPQTI 1031
>BG449141 similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (33%)
Length = 690
Score = 105 bits (261), Expect = 8e-23
Identities = 52/171 (30%), Positives = 93/171 (53%)
Frame = +3
Query: 199 TQRDLQNYSRNLKELIKDSDADMFIDNFRRKREINPSFFYDYEADNEGKLKHVFWADGIC 258
T++D++N ++ ++L + + + R ++ +P+F ++Y D +L+++ W+
Sbjct: 174 TEKDVRNLLQSFRKLDPEEETLDLLRMCRNIKDKDPNFKFEYTLDANNRLENIAWSYASS 353
Query: 259 RKNYSLFGDVVSFDTTYRTNKYFMIFAPFTGINHHRQSITFGAALLKNEKEESFVWLFET 318
+ Y +FGD V FDTT+R + M + GIN++ FG LL++E SF W +
Sbjct: 354 IQLYDIFGDAVVFDTTHRLTAFDMPLGLWVGINNYGMPCFFGCVLLRDETVRSFSWAIKA 533
Query: 319 FLKAMGGHKPVMIITDQDGGMKNAIGAVLKGSSHRFCMWHILKKLSEKVGS 369
FL M G P I+TDQ+ +K A+ A + + H FC+W I+ K V +
Sbjct: 534 FLGFMNGKAPQTILTDQNICLKEALSAEMPMTKHAFCIWMIVAKFPSWVNA 686
>TC89977 similar to GP|5764395|gb|AAD51282.1| far-red impaired response
protein {Arabidopsis thaliana}, partial (35%)
Length = 1523
Score = 100 bits (248), Expect = 3e-21
Identities = 60/166 (36%), Positives = 85/166 (51%), Gaps = 3/166 (1%)
Frame = +2
Query: 439 TTSRSESENSFFGNYLNHNLTLVEFWVRFDSALAAQRHKELFADNNTLHSNPELNMHMNL 498
T RSES NSFF Y++ +TL EF ++ L + +E AD +TLH P L
Sbjct: 14 TAQRSESMNSFFDKYIHKKITLKEFVKQYRLILLNRYDEEEIADFDTLHKQPALKSPSPW 193
Query: 499 EKHAREVYTHENFYIFQKELWSACVDCGIEGTKEEGENLSFSILDNAVRKHREVVYC--- 555
EK +YTH F FQ E+ G + E G+ + + K E +
Sbjct: 194 EKQMSTIYTHAIFKKFQVEVLGV---AGCQSRIEVGDGTAVRYIVQDYEKDEEFLVTWKE 364
Query: 556 LSNNIAHCSCKMFESEGIPCRHILFILKGKGFSEIPSHYIVNRWTK 601
LS+ ++ C C++FE +G CRH L +L+ G S +PSHYI+ RWTK
Sbjct: 365 LSSEVS-CFCRLFEYKGFLCRHALSVLQRCGCSSVPSHYIMKRWTK 499
>CB892328 GP|15075873|em PROBABLE GLYCOSYL HYDROLASE PROTEIN {Sinorhizobium
meliloti}, partial (1%)
Length = 828
Score = 93.2 bits (230), Expect = 3e-19
Identities = 63/233 (27%), Positives = 113/233 (48%), Gaps = 5/233 (2%)
Frame = +1
Query: 29 PACEDELKPV-IGKVFDTLVEGGDFYKAYAYVAGFSVRNSIKT---KDKDGVKWKYFLCS 84
P DE + + +G+V ++ E D Y+ +A+ GFSVR + +K + K F CS
Sbjct: 127 PIETDEPRDIYLGQVVNSKDEAYDLYQEHAFKTGFSVRKGKELYYDNEKKKTRLKDFYCS 306
Query: 85 KEGFKEEKKVDKPQLLIAENSLSKSRKRKLTREGCKARLVLKRTIDGKYEVSNFYEGHSH 144
K+GFK E + KR +R C A + T +G ++V+ H+H
Sbjct: 307 KQGFKNN-----------EPDGEVAYKRADSRTNCLAMVRFNVTKEGVWKVTKLILDHNH 453
Query: 145 GLVSPSKRQFLRSARNVTSVHKNILFSCNRANVGTSKSYQIMKEQVGSYENIGCTQRDLQ 204
V +R LRS RN++++ ++ + S + + + ++VG + G +D+
Sbjct: 454 EFVPLQQRYLLRSMRNMSNLKEDRIKSLVNDAIRVTNVGGYLGKEVGGVDKRGVMLKDMH 633
Query: 205 NY-SRNLKELIKDSDADMFIDNFRRKREINPSFFYDYEADNEGKLKHVFWADG 256
NY S + I+ DA +++ + K+ + F+Y + D+E +L +VFW DG
Sbjct: 634 NYVSTGKLKFIEAGDAQSLLNHLQSKQAQDSMFYYSVQLDHESRLNNVFWRDG 792
>BQ147857 weakly similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (26%)
Length = 560
Score = 75.9 bits (185), Expect = 5e-14
Identities = 42/132 (31%), Positives = 67/132 (49%), Gaps = 4/132 (3%)
Frame = +1
Query: 328 PVMIITDQDGGMKNAIGAVLKGSSHRFCMWHILKKLSE----KVGSSMDENSGFNDRFKS 383
P ++TD + +K AI + S H FC+WHIL K S+ +GS DE R
Sbjct: 4 PQTLLTDHNTWLKEAIAVEMPESKHAFCIWHILSKFSDWXYLLLGSQYDEWKADFHR--- 174
Query: 384 CVWNSESSEEFDLEWNNIISDYSLEGNGWLSTMYDLRSMWIPAYFKDTFMAGILRTTSRS 443
++N E E+F+ W ++ Y L N + ++Y LR+ W + + F AG L + ++
Sbjct: 175 -LYNLEMXEDFEKSWRQMVDKYGLHANKHIISLYSLRTFWALPFLRRYFFAG-LTSXCQT 348
Query: 444 ESENSFFGNYLN 455
ES N F +L+
Sbjct: 349 ESINVFIQRFLS 384
>BE124675 weakly similar to GP|13365573|db hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (4%)
Length = 448
Score = 70.1 bits (170), Expect = 3e-12
Identities = 39/135 (28%), Positives = 64/135 (46%)
Frame = +2
Query: 467 FDSALAAQRHKELFADNNTLHSNPELNMHMNLEKHAREVYTHENFYIFQKELWSACVDCG 526
F+ + QR+KE+ A + P+L ++ + KHA YT F +FQ+ + ++
Sbjct: 2 FERIVEEQRYKEIEASDEMKGCLPKLMGNVVVLKHASVAYTPRAFEVFQQR-YEKSLNVI 178
Query: 527 IEGTKEEGENLSFSILDNAVRKHREVVYCLSNNIAHCSCKMFESEGIPCRHILFILKGKG 586
+ K +G + + + V + S+N CSC FE G C H L +L +
Sbjct: 179 VNQHKRDGYLFEYKVNTYGHARQYTVTFSSSDNTVVCSCMKFEHVGFLCSHALKVLDNRN 358
Query: 587 FSEIPSHYIVNRWTK 601
+PS YI+ RWTK
Sbjct: 359 IKVVPSRYILKRWTK 403
>CB893278 weakly similar to PIR|T05645|T056 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (5%)
Length = 821
Score = 67.8 bits (164), Expect = 1e-11
Identities = 44/141 (31%), Positives = 75/141 (52%), Gaps = 2/141 (1%)
Frame = +3
Query: 25 VEWIPACEDELKPVIGKVFDTLVEGGDFYKAYAYVAGFSVR--NSIKTKDKDGVKWKYFL 82
VE A D L+P IG F++ E FY Y GF+VR ++ +++ + + + F+
Sbjct: 279 VEQPVADSDTLEPFIGMQFNSREEARGFYDGYGRRIGFTVRIHHNRRSRVNNELIGQDFV 458
Query: 83 CSKEGFKEEKKVDKPQLLIAENSLSKSRKRKLTREGCKARLVLKRTIDGKYEVSNFYEGH 142
CSKEGF+ +K V + ++ TREGC+A + L +GK+ V+ F + H
Sbjct: 459 CSKEGFRAKKYVHRKDRVLPPPPA--------TREGCQAMIRLALRDEGKWVVTKFVKEH 614
Query: 143 SHGLVSPSKRQFLRSARNVTS 163
+H L+SP + + S +++ S
Sbjct: 615 THKLMSPGEVPWRGSGKHLVS 677
>BE239532
Length = 645
Score = 63.5 bits (153), Expect = 3e-10
Identities = 39/107 (36%), Positives = 53/107 (49%), Gaps = 1/107 (0%)
Frame = +3
Query: 500 KHAREVYTHENFYIFQKE-LWSACVDCGIEGTKEEGENLSFSILDNAVRKHREVVYCLSN 558
+HA VYTH+ F IF E L IE ++ + +L++ K V + SN
Sbjct: 48 RHASTVYTHKGFRIFLNEYLEGTGGSTSIEISQSNNLSNHEVMLNHKPNKKYAVTFDSSN 227
Query: 559 NIAHCSCKMFESEGIPCRHILFILKGKGFSEIPSHYIVNRWTKLATS 605
+CSC+ F+S GI C H L I KG IP Y + RW+K A S
Sbjct: 228 MKINCSCRKFDSMGILCSHALRIYNIKGILRIPDQYFLKRWSKNARS 368
>CA917189 similar to PIR|T05645|T05 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (12%)
Length = 312
Score = 63.2 bits (152), Expect = 4e-10
Identities = 31/85 (36%), Positives = 46/85 (53%)
Frame = +1
Query: 320 LKAMGGHKPVMIITDQDGGMKNAIGAVLKGSSHRFCMWHILKKLSEKVGSSMDENSGFND 379
L AM G P+ I TD D +++AI V + HRFC WHI K+ EK+ + F
Sbjct: 1 LTAMSGRPPLSITTDHDSVIQSAIMQVFPDTRHRFCKWHIFKQCQEKLSHIFLQFPNFEA 180
Query: 380 RFKSCVWNSESSEEFDLEWNNIISD 404
F CV ++S +EF+ W++ +D
Sbjct: 181 EFHKCVNLTDSIDEFESCWSHF*TD 255
>TC82323 similar to GP|7673677|gb|AAF66982.1| transposase {Zea mays},
partial (2%)
Length = 1165
Score = 57.4 bits (137), Expect = 2e-08
Identities = 54/200 (27%), Positives = 79/200 (39%), Gaps = 12/200 (6%)
Frame = +1
Query: 553 VYCLSNNIAHCSCKMFESEGIPCRHILFILKGKGFSEIPSHYIVNRWTKLA-----TSKP 607
VY ++ CSC++FES GIPC H+ F++K + IP ++ RWTK A S
Sbjct: 1 VYNAASLEFQCSCRLFESRGIPCCHVFFVMKEEDVDHIPKCLVMTRWTKNAKGEFLNSDS 180
Query: 608 IFDCDGNVLEA------CSKFESESTLVSKTWSQLLKCMHMAGTNKEKLLLIYDEGCSIE 661
+ D N++E CS F + SK M +++L + + C +E
Sbjct: 181 NGEIDANMIELARFGAYCSAFTTFCKEASKKNGVFRDIM-------DEILNLQKKYCDVE 339
Query: 662 -LKMSKMKTDVVSRPLDELEAFIGSNVAKTIEVLPPEPSNTKGCGKRIKGGKEKAMEQQQ 720
SK TD +V P +KG K+ K
Sbjct: 340 DSTRSKSFTD--------------------DQVGDPVTVKSKGAPKKKKNA--------A 435
Query: 721 KRTRLCKACKQYASHDSRNC 740
K R C CK SH +R+C
Sbjct: 436 KSVRHCSRCKS-TSHTARHC 492
>TC90562 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20
{Arabidopsis thaliana}, partial (66%)
Length = 1169
Score = 56.6 bits (135), Expect = 3e-08
Identities = 38/123 (30%), Positives = 64/123 (51%), Gaps = 3/123 (2%)
Frame = +2
Query: 36 KPVIGKVFDTLVEGGDFYKAYAYVAGFSVRNS-IKTKDKDG-VKWKYFLCSKEGFKEEKK 93
+P IG+ F + E FY +YA GF +R S + +DG V + +C+KEGF+ K
Sbjct: 320 EPYIGQEFVSEAEAHAFYNSYATRVGFVIRVSKLSRSRRDGSVIGRALVCNKEGFRMPDK 499
Query: 94 VDKPQLLIAENSLSKSRKRKLTREGCKARLVLKRTIDGKYEVSNFYEGHSHGLV-SPSKR 152
+K R+R TR GC+A +++++ G + ++ F + H+H L S+R
Sbjct: 500 REKIV-----------RQRAETRVGCRAMIMVRKLNSGLWSITKFVKEHTHPLTPGKSRR 646
Query: 153 QFL 155
F+
Sbjct: 647 DFV 655
>TC92024 similar to PIR|T05645|T05645 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (11%)
Length = 938
Score = 55.5 bits (132), Expect = 8e-08
Identities = 24/45 (53%), Positives = 30/45 (66%)
Frame = +3
Query: 561 AHCSCKMFESEGIPCRHILFILKGKGFSEIPSHYIVNRWTKLATS 605
A CSC+MFE G+ CRHIL + + +PSHYI+ RWTK A S
Sbjct: 87 ATCSCQMFEFSGLLCRHILAVFRVTNVLTLPSHYILKRWTKNAKS 221
>CA922976 weakly similar to GP|9502168|gb|A contains simlarity to Arabidopsis
thaliana far-red impaired response protein
(GB:AAD51282.1), partial (11%)
Length = 827
Score = 48.5 bits (114), Expect = 9e-06
Identities = 22/50 (44%), Positives = 29/50 (58%)
Frame = -3
Query: 550 REVVYCLSNNIAHCSCKMFESEGIPCRHILFILKGKGFSEIPSHYIVNRW 599
R V++ CSCK FES GI CRH L IL K + ++P Y ++RW
Sbjct: 741 RHVLWLPEKEQILCSCKEFESSGILCRHALRILVTKNYFQLPEKYYLSRW 592
>TC81129 similar to GP|5764395|gb|AAD51282.1| far-red impaired response
protein {Arabidopsis thaliana}, partial (13%)
Length = 721
Score = 45.4 bits (106), Expect = 8e-05
Identities = 31/112 (27%), Positives = 52/112 (45%), Gaps = 2/112 (1%)
Frame = +2
Query: 32 EDELKPVIGKVFDTLVEGGDFYKAYAYVAGF--SVRNSIKTKDKDGVKWKYFLCSKEGFK 89
+ + +P G F+T FY+ YA GF S++NS ++K F CS+ G
Sbjct: 398 DTDFEPPNGIEFETHEAAYSFYQEYAKSMGFTTSIKNSRRSKKTKEFIDAKFACSRYG-- 571
Query: 90 EEKKVDKPQLLIAENSLSKSRKRKLTREGCKARLVLKRTIDGKYEVSNFYEG 141
+ E+ S++ + + CKA + +KR DGK+ + FY+G
Sbjct: 572 ----------VTPESDGRSSQRSSVKKTDCKACMHVKRNPDGKWNIFEFYKG 697
>TC79804 weakly similar to GP|6175165|gb|AAF04891.1| Mutator-like
transposase {Arabidopsis thaliana}, partial (57%)
Length = 2129
Score = 42.4 bits (98), Expect = 7e-04
Identities = 51/217 (23%), Positives = 82/217 (37%), Gaps = 19/217 (8%)
Frame = +1
Query: 234 PSFFYDYEADNEGKLKHVFW--ADGICRKNY-----SLFG------DVVSFDTTYRTNKY 280
PSF + + N G + VF AD ++ + S+ G +V+ +KY
Sbjct: 616 PSFCEEIKKTNPGSVAKVFTTGADSRFQRLFISFYASIHGFVNGCLPIVALGGIQLKSKY 795
Query: 281 FMIFAPFTGINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGHKPVM----IITDQD 336
F T + A++ E +ES+ W A+ + M ++D
Sbjct: 796 LSTFLSATSFDADGGLFPLAFAVVDVENDESWTWFLSELHNALEVNTECMPQIIFLSDGQ 975
Query: 337 GGMKNAIGAVLKGSSHRFCMWHILKKLSEKVGSSMDENSGFNDRFKSCVWNSESSEEFDL 396
G+ +AI SSH FCM H LSE +G N R +W++ + +
Sbjct: 976 KGIVDAIRRKFPRSSHAFCMRH----LSENIGKEFK-----NSRLIHLLWSAAYATTINA 1128
Query: 397 --EWNNIISDYSLEGNGWLSTMYDLRSMWIPAYFKDT 431
E I + S + WL + S W YF+ T
Sbjct: 1129FREKMAEIEEVSPNASMWLQHFHP--SQWALVYFEGT 1233
>TC81450 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20
{Arabidopsis thaliana}, partial (59%)
Length = 832
Score = 42.0 bits (97), Expect = 9e-04
Identities = 36/147 (24%), Positives = 68/147 (45%), Gaps = 6/147 (4%)
Frame = +2
Query: 73 KDGVK-WKYFLCSKEGFKEEKKVDKPQLLIAENSLSKSRKRKLTREGCKARLVLKRTIDG 131
KDG + +C+KEG++ K +K R+R TR GC+A +++++ G
Sbjct: 149 KDGTAIGRALVCNKEGYRMPDKREKIV-----------RQRAETRVGCRAMIMMRKINSG 295
Query: 132 KYEVSNFYEGHSHGLVSPSKRQFLRSARNVTSVHKNILFSCNRANVGTSKS--YQIMKEQ 189
K+ ++ F + H+H L S R+ + + H I + + +S Y+ E
Sbjct: 296 KWVITKFVKEHTHPLNPGSSRRDMFEIYPSQNEHDKIRELSQQLAIEKKRSVTYKRQLEV 475
Query: 190 VGSY---ENIGCTQRDLQNYSRNLKEL 213
+ Y N+G + R +Q ++KE+
Sbjct: 476 IFDYIEEHNVGLS-RKMQRIVDSVKEM 553
>BF646907 similar to GP|15983442|gb| At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (4%)
Length = 621
Score = 42.0 bits (97), Expect = 9e-04
Identities = 34/97 (35%), Positives = 49/97 (50%), Gaps = 8/97 (8%)
Frame = +1
Query: 40 GKVFDTLVEGGDFYKAYAYVAGFSVR-----NSIKTKDKD---GVKWKYFLCSKEGFKEE 91
GKVF++ E +FY +A GFS+R SIK + D GV + F+C + G
Sbjct: 328 GKVFNSDDEAYNFYCLFARKNGFSIRRHHVYKSIKNQSDDNPLGVYKREFVCHRAG---T 498
Query: 92 KKVDKPQLLIAENSLSKSRKRKLTREGCKARLVLKRT 128
VDK +N + RKRK +R C A+L++ T
Sbjct: 499 ISVDK------DNEVEGKRKRKSSRCNCGAKLLVNIT 591
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.134 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,405,427
Number of Sequences: 36976
Number of extensions: 377890
Number of successful extensions: 1944
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 1912
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1931
length of query: 745
length of database: 9,014,727
effective HSP length: 103
effective length of query: 642
effective length of database: 5,206,199
effective search space: 3342379758
effective search space used: 3342379758
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148396.11


