

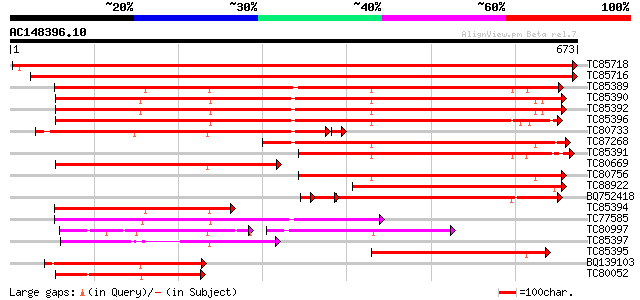
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148396.10 + phase: 0 /partial
(673 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa prot... 1258 0.0
TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chape... 1129 0.0
TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protei... 574 e-164
TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 ... 566 e-162
TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chape... 563 e-161
TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protei... 558 e-159
TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa prot... 315 1e-87
TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chape... 297 8e-81
TC85391 homologue to PIR|T06598|T06598 dnaK-type molecular chape... 290 9e-79
TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 290 1e-78
TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 280 1e-75
TC88922 homologue to SP|Q02028|HS7S_PEA Stromal 70 kDa heat shoc... 275 3e-74
BQ752418 homologue to SP|P78695|GR78 78 kDa glucose-regulated pr... 235 2e-67
TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic retic... 249 2e-66
TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock... 224 8e-59
TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [im... 143 4e-54
TC85397 homologue to GP|170386|gb|AAA99920.1|| glucose-regulated... 204 9e-53
TC85395 homologue to GP|6911553|emb|CAB72130.1 heat shock protei... 196 3e-50
BQ139103 homologue to GP|2655420|gb| heat shock cognate protein ... 195 4e-50
TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chapero... 174 1e-43
>TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa protein
mitochondrial precursor. [Kidney bean French bean]
{Phaseolus vulgaris}, complete
Length = 2371
Score = 1258 bits (3254), Expect = 0.0
Identities = 655/675 (97%), Positives = 659/675 (97%), Gaps = 5/675 (0%)
Frame = +2
Query: 4 PTQACCS-----ILIVSQTYSDVVLQLTGNTKPAYVGHNWSSLSRPFSSRAAGNDVIGID 58
P Q CS +++ S T + LTGNTKPAYVGHNWSSLSRPFSSRAAGNDVIGID
Sbjct: 98 PLQPLCSALSAAVMLASSTVT-AFRSLTGNTKPAYVGHNWSSLSRPFSSRAAGNDVIGID 274
Query: 59 LGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPENTISG 118
LGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPENTISG
Sbjct: 275 LGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPENTISG 454
Query: 119 AKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAKGQQYSPSQIGAFVLTKMKETAE 178
AKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAKGQQYSPSQIGAFVLTKMKETAE
Sbjct: 455 AKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAKGQQYSPSQIGAFVLTKMKETAE 634
Query: 179 AYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYGMNKEGLIA 238
AYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYGMNKEGLIA
Sbjct: 635 AYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYGMNKEGLIA 814
Query: 239 VFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRSESIDLSKD 298
VFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRSESIDLSKD
Sbjct: 815 VFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRSESIDLSKD 994
Query: 299 KLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEALVNNLIER 358
KLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEALVNNLIER
Sbjct: 995 KLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEALVNNLIER 1174
Query: 359 TKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIFGKSPCKGVNPDEAVAMGA 418
TKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIFGKSPCKGVNPDEAVAMGA
Sbjct: 1175TKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIFGKSPCKGVNPDEAVAMGA 1354
Query: 419 ALQGGILRGDVKELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQ 478
ALQGGILRGDVKELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQ
Sbjct: 1355ALQGGILRGDVKELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQ 1534
Query: 479 VGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGK 538
VGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGK
Sbjct: 1535VGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGK 1714
Query: 539 EQQITIKSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEKSLSEYRE 598
EQQITIKSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEKSLSEYRE
Sbjct: 1715EQQITIKSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEKSLSEYRE 1894
Query: 599 KIPAEVAKEIEDAVSDLRSAMAGDSADEIKSKLDAANKAVSKIGEHMSGGSSSGPSSDGS 658
KIPAEVAKEIEDAVSDLRSAMAGDSADEIKSKLDAANKAVSKIGEHMSGGSSSGPSSDGS
Sbjct: 1895KIPAEVAKEIEDAVSDLRSAMAGDSADEIKSKLDAANKAVSKIGEHMSGGSSSGPSSDGS 2074
Query: 659 QGGDQAPEAEYEEVK 673
QGGDQAPEAEYEEVK
Sbjct: 2075QGGDQAPEAEYEEVK 2119
>TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chaperone PHSP1
precursor mitochondrial - garden pea, partial (98%)
Length = 2422
Score = 1129 bits (2920), Expect = 0.0
Identities = 584/653 (89%), Positives = 617/653 (94%), Gaps = 4/653 (0%)
Frame = +1
Query: 25 LTGNTKPAYVGHNWSSLSRPFSSRAAGNDVIGIDLGTTNSCVSVMEGKNPKVVENSEGAR 84
LTG+TKP+Y W+SL+RPFSSR AG+DVIGIDLGTTNSCVS+MEGKNPKV+ENSEGAR
Sbjct: 157 LTGSTKPSYAAQKWASLARPFSSRPAGSDVIGIDLGTTNSCVSLMEGKNPKVIENSEGAR 336
Query: 85 TTPSVVAFTQKGELLVGTPAKRQAVTNPENTISGAKRLIGRRFDDPQTQKEMKMVPYKIV 144
TTPSVVAF QKGELLVGTPAKRQAVTNP NT+ G KRLIGRRFDDPQTQKEMKMVPYKIV
Sbjct: 337 TTPSVVAFNQKGELLVGTPAKRQAVTNPTNTLFGTKRLIGRRFDDPQTQKEMKMVPYKIV 516
Query: 145 KAPNGDAWVEAKGQQYSPSQIGAFVLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQAT 204
KAPNGDAWVE QQYSPSQIGAFVLTKMKETAEAYLGKT+ KAV+TVPAYFNDAQRQAT
Sbjct: 517 KAPNGDAWVEINKQQYSPSQIGAFVLTKMKETAEAYLGKTISKAVVTVPAYFNDAQRQAT 696
Query: 205 KDAGRIAGLEVLRIINEPTAAALSYGMN-KEGLIAVFDLGGGTFDVSILEISNGVFEVKA 263
KDAGRIAGLEV RIINEPTAAALSYGMN KEGLIAVFDLGGGTFDVSILEISNGVFEVKA
Sbjct: 697 KDAGRIAGLEVKRIINEPTAAALSYGMNNKEGLIAVFDLGGGTFDVSILEISNGVFEVKA 876
Query: 264 TNGDTFLGGEDFDNALLDFLVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTE 323
TNGDTFLGGEDFDNALLDFLV+EFKR++SIDL+KDKLALQRLREAAEKAKIELSSTSQTE
Sbjct: 877 TNGDTFLGGEDFDNALLDFLVSEFKRTDSIDLAKDKLALQRLREAAEKAKIELSSTSQTE 1056
Query: 324 INLPFITADASGAKHLNITLTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLV 383
INLPFITADASGAKHLNITLTRSKFEALVNNLIERTKAPC+SCLKDANIS KD+DEVLLV
Sbjct: 1057INLPFITADASGAKHLNITLTRSKFEALVNNLIERTKAPCKSCLKDANISIKDVDEVLLV 1236
Query: 384 GGMTRVPKVQEVVSTIFGKSPCKGVNPDEAVAMGAALQGGILRGDVKELLLLDVTPLSLG 443
GGMTRVPKVQEVVS IFGKSP KGVNPDEAVAMGAALQGGILRGDVKELLLLDVTPLSLG
Sbjct: 1237GGMTRVPKVQEVVSEIFGKSPSKGVNPDEAVAMGAALQGGILRGDVKELLLLDVTPLSLG 1416
Query: 444 IETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIK--VLQGEREMASDNKMLGEFE 501
IETLGGIFTRLI+RNTTIPTKKSQVFSTAADNQTQ G +G REMA+DNK LGEF+
Sbjct: 1417IETLGGIFTRLISRNTTIPTKKSQVFSTAADNQTQRGYXRCSQEGXREMAADNKSLGEFD 1596
Query: 502 LVGLPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIKSSGGLSDDEIQNMVKE 561
LVG+PPAPRG+PQIEVTFDIDANGIVTVSAKDKSTGKEQQITI+SSGGLSDDEI NMVKE
Sbjct: 1597LVGIPPAPRGLPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIRSSGGLSDDEINNMVKE 1776
Query: 562 AELHAQKDQERKSLIDIRNSADTSIYSIEKSLSEYREKIPAEVAKEIEDAVSDLRSAMAG 621
AELHAQ+DQERK+LIDI+NSADTSIYSIEKSLSEYREKIP+EVAKEIE++VSDLR+AM G
Sbjct: 1777AELHAQRDQERKALIDIKNSADTSIYSIEKSLSEYREKIPSEVAKEIENSVSDLRTAMEG 1956
Query: 622 DSADEIKSKLDAANKAVSKIGEHMSGGSSSGPSSDGSQ-GGDQAPEAEYEEVK 673
+S DEIK+KLDAANKAVSKIG+HMSGGSS G S GS GGDQAPEAEYEEVK
Sbjct: 1957ESVDEIKTKLDAANKAVSKIGQHMSGGSSGGSSDGGSPGGGDQAPEAEYEEVK 2115
>TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protein 4
precursor (BiP 4) (78 kDa glucose-regulated protein
homolog 4) (GRP 78-4)., partial (94%)
Length = 2265
Score = 574 bits (1479), Expect = e-164
Identities = 313/627 (49%), Positives = 424/627 (66%), Gaps = 23/627 (3%)
Frame = +2
Query: 54 VIGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPE 113
VIGIDLGTT SCV V + + +++ N +G R TPS V+FT E L+G AK A NPE
Sbjct: 161 VIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVSFTDD-ERLIGEAAKNLAAVNPE 337
Query: 114 NTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAKGQQ-----YSPSQIGAF 168
TI KRLIGR+F D + Q++MK+VPYKIV +G +++ + + +SP ++ A
Sbjct: 338 RTIFDVKRLIGRKFADKEVQRDMKLVPYKIVNK-DGKPYIQVRVKDGETKVFSPEEVSAM 514
Query: 169 VLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALS 228
+LTKMKETAEA+LGKT+ AV+TVPAYFNDAQRQATKDAG IAGL V RIINEPTAAA++
Sbjct: 515 ILTKMKETAEAFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAAAIA 694
Query: 229 YGMNKEG---LIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVN 285
YG++K+G I VFDLGGGTFDVSIL I NGVFEV +TNGDT LGGEDFD ++++ +
Sbjct: 695 YGLDKKGGEKNILVFDLGGGTFDVSILTIDNGVFEVLSTNGDTHLGGEDFDQRIMEYFIK 874
Query: 286 EFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTR 345
K+ S D+SKD AL +LR +E+AK LSS Q + + + ++ LTR
Sbjct: 875 LIKKKHSKDISKDNRALGKLRRESERAKRALSSQHQVRVEIESLFDGVDFSE----PLTR 1042
Query: 346 SKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIF-GKSP 404
++FE L N+L +T P + + DA + IDE++LVGG TR+PKVQ+++ F GK P
Sbjct: 1043ARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTRIPKVQQLLKDYFDGKEP 1222
Query: 405 CKGVNPDEAVAMGAALQGGILRGD----VKELLLLDVTPLSLGIETLGGIFTRLINRNTT 460
KGVNPDEAVA GAA+QG IL G+ K++LLLDV PL+LGIET+GG+ T+LI RNT
Sbjct: 1223NKGVNPDEAVAFGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTKLIPRNTV 1402
Query: 461 IPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTFD 520
IPTKKSQVF+T D QT V I+V +GER + D ++LG F+L G+PPAPRG PQIEVTF+
Sbjct: 1403IPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGNFDLSGIPPAPRGTPQIEVTFE 1582
Query: 521 IDANGIVTVSAKDKSTGKEQQITI-KSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIR 579
+DANGI+ V A+DK TGK ++ITI G LS +EI MV+EAE A++D++ K ID R
Sbjct: 1583VDANGILNVRAEDKGTGKSEKITITNEKGRLSQEEIDRMVREAEEFAEEDKKVKERIDAR 1762
Query: 580 NSADTSIYSIEKSLSE---YREKIPAEVAKEIEDAVS------DLRSAMAGDSADEIKSK 630
N+ +T +Y+++ +S+ +K+ ++ ++IE AV D + + +E +
Sbjct: 1763NALETYVYNMKNQISDKDKLADKLESDEKEKIEAAVKEALEWLDDNQTVEKEEFEEKLKE 1942
Query: 631 LDAANKAVSKIGEHMSGGSSSGPSSDG 657
++A + SGG+ G S +G
Sbjct: 1943VEAVCNPIITAVYQRSGGAPGGASGEG 2023
>TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 kDa protein
2. [Tomato] {Lycopersicon esculentum}, complete
Length = 2278
Score = 566 bits (1459), Expect = e-162
Identities = 310/632 (49%), Positives = 418/632 (66%), Gaps = 25/632 (3%)
Frame = +1
Query: 55 IGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPEN 114
IGIDLGTT SCV V + +++ N +G RTTPS VAFT E L+G AK Q NP N
Sbjct: 163 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDT-ERLIGDAAKNQVAMNPTN 339
Query: 115 TISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWV----EAKGQQYSPSQIGAFVL 170
T+ AKRLIGRRF D Q +MK+ P+K++ P + +A+ +Q+S +I + VL
Sbjct: 340 TVFDAKRLIGRRFGDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQFSAEEISSMVL 519
Query: 171 TKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYG 230
KMKE AEAYLG T+ AV+TVPAYFND+QRQATKDAG I+GL VLRIINEPTAAA++YG
Sbjct: 520 MKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVLRIINEPTAAAIAYG 699
Query: 231 MNKEGL------IAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLV 284
++K+ + +FDLGGGTFDVS+L I G+FEVKAT GDT LGGEDFDN +++ V
Sbjct: 700 LDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 879
Query: 285 NEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLT 344
EFKR D+S + AL+RLR A E+AK LSST+QT I + + T+T
Sbjct: 880 QEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGID----FYTTIT 1047
Query: 345 RSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIF-GKS 403
R++FE L +L + P + CL+DA + + +V+LVGG TR+PKVQ+++ F GK
Sbjct: 1048RARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQDFFNGKE 1227
Query: 404 PCKGVNPDEAVAMGAALQGGILRGD----VKELLLLDVTPLSLGIETLGGIFTRLINRNT 459
CK +NPDEAVA GAA+Q IL G+ V++LLLLDVTPLSLG+ET GG+ T LI RNT
Sbjct: 1228LCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNT 1407
Query: 460 TIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTF 519
TIPTKK QVFST +DNQ V I+V +GER DN +LG+FEL G+PPAPRG+PQI V F
Sbjct: 1408TIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCF 1587
Query: 520 DIDANGIVTVSAKDKSTGKEQQITI-KSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDI 578
DIDANGI+ VSA+DK+TG++ +ITI G LS +EI+ MV+EAE + +D+E K ++
Sbjct: 1588DIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDEEHKKKVEA 1767
Query: 579 RNSADTSIYSIEKSLSEYR--EKIPAEVAKEIEDAVSDLRSAMAGD---SADEIKSKL-- 631
+N+ + Y++ ++ + + K+ A+ K+IEDA+ + G+ ADE + K+
Sbjct: 1768KNALENYAYNMRNTIKDDKIASKLSADDKKKIEDAIEGAIQWLDGNQLGEADEFEDKMKE 1947
Query: 632 --DAANKAVSKIGEHMSGGSSSGPSSDGSQGG 661
N ++++ + G + DG G
Sbjct: 1948LEGICNPIIARMYQGAGGDAGGAMDEDGPTAG 2043
>TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chaperone
hsc70-3 - tomato, complete
Length = 2295
Score = 563 bits (1452), Expect = e-161
Identities = 307/632 (48%), Positives = 418/632 (65%), Gaps = 25/632 (3%)
Frame = +1
Query: 55 IGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPEN 114
IGIDLGTT SCV V + +++ N +G RTTPS VAFT E L+G AK Q NP N
Sbjct: 157 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDT-ERLIGDAAKNQVAMNPTN 333
Query: 115 TISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWV----EAKGQQYSPSQIGAFVL 170
T+ AKRLIGRR D Q +MK+ P+K++ P + +A+ +Q+S +I + VL
Sbjct: 334 TVFDAKRLIGRRISDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQFSAEEISSMVL 513
Query: 171 TKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYG 230
KMKE AEAYLG T+ AV+TVPAYFND+QRQATKDAG I+GL V+RIINEPTAAA++YG
Sbjct: 514 MKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIINEPTAAAIAYG 693
Query: 231 MNKEGL------IAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLV 284
++K+ + +FDLGGGTFDVS+L I G+FEVKAT GDT LGGEDFDN +++ V
Sbjct: 694 LDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 873
Query: 285 NEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLT 344
EFKR D+S + AL+R+R A E+AK LSST+QT I + + T+T
Sbjct: 874 QEFKRKNKKDISGNPRALRRVRTACERAKRTLSSTAQTTIEIDSLFEGID----FYTTIT 1041
Query: 345 RSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIF-GKS 403
R++FE L +L + P + CL+DA + +D+V+LVGG TR+PKVQ+++ F GK
Sbjct: 1042RARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVDDVVLVGGSTRIPKVQQLLQDFFNGKE 1221
Query: 404 PCKGVNPDEAVAMGAALQGGILRGD----VKELLLLDVTPLSLGIETLGGIFTRLINRNT 459
CK +NPDEAVA GAA+Q IL G+ V++LLLLDVTPLSLG+ET GG+ T LI RNT
Sbjct: 1222LCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNT 1401
Query: 460 TIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTF 519
TIPTKK QVFST +DNQ V I+V +GER DN +LG+FEL G+PPAPRG+PQI V F
Sbjct: 1402TIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCF 1581
Query: 520 DIDANGIVTVSAKDKSTGKEQQITI-KSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDI 578
DIDANGI+ VSA+DK+TG++ +ITI G LS ++I+ MV+EAE + +D+E K ++
Sbjct: 1582DIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEHKRKVEA 1761
Query: 579 RNSADTSIYSIEKSLSEYR--EKIPAEVAKEIEDAVSDLRSAMAGD---SADEIKSKL-- 631
+N+ + Y++ ++ + + K+ A+ K+IEDA+ + G+ ADE + K+
Sbjct: 1762KNALENYAYNMRNTIKDDKIASKLSADDKKKIEDAIEGAIQWLDGNQLGEADEFEDKMKE 1941
Query: 632 --DAANKAVSKIGEHMSGGSSSGPSSDGSQGG 661
N ++++ + G + DG G
Sbjct: 1942LEGICNPIIARMYQGAGGDAGGAMDEDGPAAG 2037
>TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protein 70
{Cucumis sativus}, complete
Length = 2320
Score = 558 bits (1438), Expect = e-159
Identities = 311/629 (49%), Positives = 415/629 (65%), Gaps = 27/629 (4%)
Frame = +2
Query: 55 IGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPEN 114
IGIDLGTT SCV V + +++ N +G RTTPS VAFT E L+G AK Q NP N
Sbjct: 134 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDS-ERLIGDAAKNQVAMNPTN 310
Query: 115 TISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAW--VEAKGQQ--YSPSQIGAFVL 170
T+ AKRLIGRR D Q +MK+ P+K++ P V KG++ ++ +I + VL
Sbjct: 311 TVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEEKLFASEEISSMVL 490
Query: 171 TKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYG 230
KM+E AEAYLG T+ AV+TVPAYFND+QRQATKDAG IAGL VLRIINEPTAAA++YG
Sbjct: 491 IKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYG 670
Query: 231 MNKEGL------IAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLV 284
++K+ + +FDLGGGTFDVS+L I G+FEVKAT GDT LGGEDFDN ++ V
Sbjct: 671 LDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVTHFV 850
Query: 285 NEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLT 344
EFKR D+S + AL+RLR A E+AK LSST+QT I + + T+T
Sbjct: 851 QEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVD----FYTTIT 1018
Query: 345 RSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIF-GKS 403
R++FE L +L + P + CL+DA + + +V+LVGG TR+PKVQ+++ F GK
Sbjct: 1019RARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLLQDFFNGKE 1198
Query: 404 PCKGVNPDEAVAMGAALQGGILRGD----VKELLLLDVTPLSLGIETLGGIFTRLINRNT 459
CK +NPDEAVA GAA+Q IL G+ V++LLLLDVTPLS G+ET GG+ T LI RNT
Sbjct: 1199LCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSQGLETAGGVMTVLIPRNT 1378
Query: 460 TIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTF 519
TIPTKK QVFST +DNQ V I+V +GER DN +LG+FEL G+PPAPRG+PQI V F
Sbjct: 1379TIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTKDNNLLGKFELSGIPPAPRGVPQITVCF 1558
Query: 520 DIDANGIVTVSAKDKSTGKEQQITI-KSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDI 578
DIDANGI+ VSA+DK+TG++ +ITI G LS +EI+ MV+EAE + +D+E K ++
Sbjct: 1559DIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDEEHKKKVEA 1738
Query: 579 RNSADTSIYSIEKSLSEYREKIPAEVA----KEIEDAVSDL-------RSAMAGDSADEI 627
+NS + Y++ ++ + EKI ++++ K+IEDA+ + A A + D++
Sbjct: 1739KNSLENYAYNMRNTIKD--EKISSKLSGGDKKQIEDAIEGAIQWLDANQLAEADEFEDKM 1912
Query: 628 KSKLDAANKAVSKIGEHMSGGSSSGPSSD 656
K N ++K+ GG+ GP D
Sbjct: 1913KELETICNPIIAKM---YQGGAGEGPEVD 1990
>TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa protein
(HSP70). {Neurospora crassa}, partial (54%)
Length = 1258
Score = 315 bits (808), Expect(2) = 1e-87
Identities = 176/358 (49%), Positives = 228/358 (63%), Gaps = 7/358 (1%)
Frame = +2
Query: 31 PAYVGHNWSSLSRPFSSRAAGNDVIGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVV 90
P YV + +S+ + A + IGIDLGTT SCV +++ N +G RTTPS V
Sbjct: 137 PRYVTYTHTSI------KMATSFAIGIDLGTTYSCVGRYANDKIEIIANDQGNRTTPSYV 298
Query: 91 AFTQKGELLVGTPAKRQAVTNPENTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKA---P 147
AF E L+G AK Q NP NT+ AKRLIGR+F D + Q +MK P+K++ P
Sbjct: 299 AFNDT-ERLIGDAAKNQVAMNPHNTVFDAKRLIGRKFSDSEVQADMKHFPFKVIDKGGKP 475
Query: 148 NGDAWVEAKGQQYSPSQIGAFVLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDA 207
N + + + + ++P +I A VL KM+ETAEAYLG V AVITVPAYFND+QRQATKDA
Sbjct: 476 NIEVEFKGENKTFTPEEISAMVLVKMRETAEAYLGGQVTNAVITVPAYFNDSQRQATKDA 655
Query: 208 GRIAGLEVLRIINEPTAAALSYGMNK----EGLIAVFDLGGGTFDVSILEISNGVFEVKA 263
G IAGL VLRIINEPTAAA++YG++K E + +FDLGGGTFDVS+L I G+FEVK+
Sbjct: 656 GLIAGLNVLRIINEPTAAAIAYGLDKKAEGERNVLIFDLGGGTFDVSLLTIEEGIFEVKS 835
Query: 264 TNGDTFLGGEDFDNALLDFLVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTE 323
T GDT LGGEDFDN L++ VNEFKR DLS + AL+RLR A E+AK LSS++QT
Sbjct: 836 TAGDTHLGGEDFDNRLVNHFVNEFKRKHKKDLSSNARALRRLRTACERAKRTLSSSAQTS 1015
Query: 324 INLPFITADASGAKHLNITLTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVL 381
I + + ++TR++FE L +L T P L DA I + E++
Sbjct: 1016IEIDSLFEGID----FYTSITRARFEELCQDLFRSTIQPVDRVLSDAKIDKSQVHEIV 1177
Score = 26.9 bits (58), Expect(2) = 1e-87
Identities = 9/18 (50%), Positives = 15/18 (83%)
Frame = +3
Query: 383 VGGMTRVPKVQEVVSTIF 400
VGG TR+P++Q+++S F
Sbjct: 1182 VGGSTRIPRIQKLISDYF 1235
>TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chaperone
HSC71.0 - garden pea, partial (59%)
Length = 1336
Score = 297 bits (761), Expect = 8e-81
Identities = 169/376 (44%), Positives = 239/376 (62%), Gaps = 11/376 (2%)
Frame = +2
Query: 301 ALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEALVNNLIERTK 360
AL+RLR A E+AK LSST+QT I + + +TR++FE L +L +
Sbjct: 5 ALRRLRTACERAKRTLSSTAQTTIEIDSLFEGID----FYSPITRARFEELNMDLFRKCM 172
Query: 361 APCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIF-GKSPCKGVNPDEAVAMGAA 419
P + CL+DA + K + +V+LVGG TR+PKVQ+++ F GK CK +NPDEAVA GAA
Sbjct: 173 EPVEKCLRDAKMDKKSVHDVVLVGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAA 352
Query: 420 LQGGILRGD----VKELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADN 475
+Q IL G+ V++LLLLDVTPLSLG+ET GG+ T LI RNTTIPTKK QVFST +DN
Sbjct: 353 VQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDN 532
Query: 476 QTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTVSAKDKS 535
Q V I+V +GER DN +LG+FEL G+PPAPRG+PQI V FDIDANGI+ VSA+DK+
Sbjct: 533 QPGVLIQVFEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKT 712
Query: 536 TGKEQQITI-KSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEKSLS 594
TG++ +ITI G LS ++I+ MV+EAE + +D+E K ++ +N+ + Y++ ++
Sbjct: 713 TGQKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEHKKKVEAKNALENYAYNMRNTIK 892
Query: 595 EYR--EKIPAEVAKEIEDAVSDLRSAMAGD---SADEIKSKLDAANKAVSKIGEHMSGGS 649
+ + K+ ++ K+IED + + + ADE + K+ + I M G
Sbjct: 893 DEKIAGKLDSDDKKKIEDTIEAAIQWLDANQLAEADEFEDKMKELEGVCNPIIAKMYQG- 1069
Query: 650 SSGPSSDGSQGGDQAP 665
+GP + G D AP
Sbjct: 1070GAGPDMGAAPGDDDAP 1117
>TC85391 homologue to PIR|T06598|T06598 dnaK-type molecular chaperone BiP-A
- soybean, partial (52%)
Length = 1231
Score = 290 bits (743), Expect = 9e-79
Identities = 159/344 (46%), Positives = 229/344 (66%), Gaps = 16/344 (4%)
Frame = +1
Query: 343 LTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIF-G 401
LTR++FE L N+L +T P + + DA + IDE++LVGG TR+PKVQ+++ F G
Sbjct: 25 LTRARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTRIPKVQQLLKDYFDG 204
Query: 402 KSPCKGVNPDEAVAMGAALQGGILRGD----VKELLLLDVTPLSLGIETLGGIFTRLINR 457
K P KGVNPDEAVA GAA+QG IL G+ K++LLLDV PL+LGIET+GG+ T+LI R
Sbjct: 205 KEPNKGVNPDEAVAFGAAVQGSILSGEGGAETKDILLLDVAPLTLGIETVGGVMTKLIPR 384
Query: 458 NTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEV 517
NT IPTKKSQVF+T D QT V I+V +GER + D ++LG+F+L G+PPAPRG PQIEV
Sbjct: 385 NTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLSGIPPAPRGTPQIEV 564
Query: 518 TFDIDANGIVTVSAKDKSTGKEQQITI-KSSGGLSDDEIQNMVKEAELHAQKDQERKSLI 576
TF++DANGI+ V A+DK TGK ++ITI G LS +EI+ MV+EAE A++D++ K I
Sbjct: 565 TFEVDANGILNVKAEDKGTGKSEKITITNEKGRLSQEEIERMVREAEEFAEEDKKVKERI 744
Query: 577 DIRNSADTSIYSIEKSLSE---YREKIPAEVAKEIEDAV-------SDLRSAMAGDSADE 626
D RN+ +T +Y+++ +S+ +K+ ++ ++IE AV D +S + ++
Sbjct: 745 DARNALETYVYNMKNQVSDKDKLADKLESDEKEKIETAVKEALEWLDDNQSVEKEEYEEK 924
Query: 627 IKSKLDAANKAVSKIGEHMSGGSSSGPSSDGSQGGDQAPEAEYE 670
+K N ++ + + SGG+ G G+ G D+ E+ E
Sbjct: 925 LKEVEAVCNPIITAVYQR-SGGAPGG----GASGEDEEDESHDE 1041
>TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (44%)
Length = 955
Score = 290 bits (742), Expect = 1e-78
Identities = 155/278 (55%), Positives = 196/278 (69%), Gaps = 10/278 (3%)
Frame = +2
Query: 55 IGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPEN 114
IGIDLGTT SCV V + +++ N +G RTTPS VAFT E L+G AK Q NP+N
Sbjct: 122 IGIDLGTTYSCVGVWQNDRVEIIPNDQGNRTTPSYVAFTDT-ERLIGDAAKNQVAMNPQN 298
Query: 115 TISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAW--VEAKGQQ--YSPSQIGAFVL 170
T+ AKRLIGRRF D Q +MK+ P+K+V P V KG++ ++ +I + VL
Sbjct: 299 TVFDAKRLIGRRFSDESVQNDMKLWPFKVVPGPAEKPMIVVNYKGEEKKFAAEEISSMVL 478
Query: 171 TKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYG 230
KM+E AEA+LG V AV+TVPAYFND+QRQATKDAG I+GL VLRIINEPTAAA++YG
Sbjct: 479 IKMREVAEAFLGHPVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEPTAAAIAYG 658
Query: 231 MNK------EGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLV 284
++K E + +FDLGGGTFDVS+L I G+FEVKAT GDT LGGEDFDN +++ V
Sbjct: 659 LDKKASRKGEQNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 838
Query: 285 NEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQT 322
+EF+R D+S + AL+RLR A E+AK LSST+QT
Sbjct: 839 SEFRRKNKKDISGNARALRRLRTACERAKRTLSSTAQT 952
>TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (53%)
Length = 1189
Score = 280 bits (716), Expect = 1e-75
Identities = 156/332 (46%), Positives = 213/332 (63%), Gaps = 13/332 (3%)
Frame = +3
Query: 343 LTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIF-G 401
+TR++FE L +L + P + CL+DA I + EV+LVGG TR+PKVQ+++ F G
Sbjct: 3 ITRARFEELNMDLFRKCMEPVEKCLRDAKIDKSHVHEVVLVGGSTRIPKVQQLLQDFFNG 182
Query: 402 KSPCKGVNPDEAVAMGAALQGGILRGD----VKELLLLDVTPLSLGIETLGGIFTRLINR 457
K CK +NPDEAVA GAA+Q IL G+ V++LLLLDVTPLSLG+ET GG+ T LI R
Sbjct: 183 KELCKSINPDEAVAYGAAVQAAILSGEGDEKVQDLLLLDVTPLSLGLETAGGVMTTLIPR 362
Query: 458 NTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEV 517
NTTIPTKK Q+FST +DNQ V I+V +GER DN +LG+FEL G+PPAPRG+PQI V
Sbjct: 363 NTTIPTKKEQIFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELTGIPPAPRGVPQINV 542
Query: 518 TFDIDANGIVTVSAKDKSTGKEQQITI-KSSGGLSDDEIQNMVKEAELHAQKDQERKSLI 576
FDIDANGI+ VSA+DK+ G + +ITI G LS +EI+ MVK+AE + +D+E K +
Sbjct: 543 CFDIDANGILNVSAEDKTAGVKNKITITNDKGRLSKEEIEKMVKDAEKYKAEDEEVKKKV 722
Query: 577 DIRNSADTSIYSIEKSLSEYR--EKIPAEVAKEIEDAVSDLRSAMAGD---SADEIKSKL 631
+ +NS + Y++ ++ + + K+ E ++IE AV D + G+ DE + K
Sbjct: 723 EAKNSIENYAYNMRNTIKDEKIGGKLSHEDKEKIEKAVEDAIQWLEGNQMAEVDEFEDKQ 902
Query: 632 DAANKAVSKIGEHMSGGSSSG--PSSDGSQGG 661
+ I M G + G P DG GG
Sbjct: 903 KELEGICNPIIAKMYQGGAGGDVPMGDGMPGG 998
>TC88922 homologue to SP|Q02028|HS7S_PEA Stromal 70 kDa heat shock-related
protein chloroplast precursor. [Garden pea] {Pisum
sativum}, partial (42%)
Length = 1092
Score = 275 bits (704), Expect = 3e-74
Identities = 135/258 (52%), Positives = 186/258 (71%), Gaps = 4/258 (1%)
Frame = +2
Query: 408 VNPDEAVAMGAALQGGILRGDVKELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQ 467
VNPDE VA+GAA+Q G+L GDV +++LLDV+PLSLG+ETLGG+ T++I RNTT+PT KS+
Sbjct: 50 VNPDEVVALGAAVQAGVLAGDVSDIVLLDVSPLSLGLETLGGVMTKIIPRNTTLPTSKSE 229
Query: 468 VFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIV 527
VFSTAAD QT V I VLQGERE DNK LG F L G+PPAPRG+PQIEV FDIDANGI+
Sbjct: 230 VFSTAADGQTSVEINVLQGEREFVRDNKSLGSFRLDGIPPAPRGVPQIEVKFDIDANGIL 409
Query: 528 TVSAKDKSTGKEQQITIKSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIY 587
+V+A DK TGK+Q ITI + L DE++ MV EAE +++D+E++ ID +N AD+ +Y
Sbjct: 410 SVAAIDKGTGKKQDITITGASTLPGDEVERMVNEAERFSKEDKEKRDAIDTKNQADSVVY 589
Query: 588 SIEKSLSEYREKIPAEVAKEIEDAVSDLRSAMAGDSADEIKSKLDAANKAVSKIGEHM-- 645
EK L E EK+P V +++E + +L+ A++G S +K + A N+ V ++G+ +
Sbjct: 590 QTEKQLKELGEKVPGPVKEKVEAKLVELKDAISGGSTQTMKDAMAALNQEVMQLGQSLYN 769
Query: 646 --SGGSSSGPSSDGSQGG 661
++GP+ GS+ G
Sbjct: 770 QPGAADAAGPTPPGSESG 823
>BQ752418 homologue to SP|P78695|GR78 78 kDa glucose-regulated protein
homolog precursor (GRP 78), partial (47%)
Length = 1032
Score = 235 bits (600), Expect(3) = 2e-67
Identities = 133/286 (46%), Positives = 185/286 (64%), Gaps = 18/286 (6%)
Frame = -1
Query: 389 VPKVQEVVSTIFG-KSPCKGVNPDEAVAMGAALQGGILRGD--VKELLLLDVTPLSLGIE 445
+PKVQ ++ FG K KG+NPDEAVA GAA+Q G+L G+ +E++L+DV PL+LGIE
Sbjct: 900 IPKVQSLIEEYFGGKKASKGINPDEAVAFGAAVQAGVLSGEEGTEEIVLMDVNPLTLGIE 721
Query: 446 TLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGL 505
T GG+ T+LI RNT IPT+KSQ+FSTAADNQ V I+V +GER + DN LG+FEL G+
Sbjct: 720 TTGGVMTKLITRNTPIPTRKSQIFSTAADNQPVVLIQVFEGERSLTKDNNQLGKFELTGI 541
Query: 506 PPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITI-KSSGGLSDDEIQNMVKEAEL 564
PPAPRG+PQIEV+F++DANGI+ VSA DK TGK++ ITI G L+ +EI MV+EAE
Sbjct: 540 PPAPRGVPQIEVSFELDANGILKVSAHDKGTGKQESITITNDKGRLTKEEIDRMVEEAEK 361
Query: 565 HAQKDQERKSLIDIRNSADTSIYSIEKSLS-----------EYREKIPAEVAKEIEDAVS 613
+A++D+ + I+ RN + +S++ ++ E +E I E KE D +
Sbjct: 360 YAEEDKATRERIEARNGLENYAFSLKNQVNDEEGLGGKIDDEDKETI-LEAVKETNDWLE 184
Query: 614 DLRSAMAGDSADEIKSKL-DAANKAVSKI--GEHMSGGSSSGPSSD 656
+ + + +E K KL + A SK+ G +GG PS D
Sbjct: 183 ENGATANTEDFEEQKEKLSNVAYPITSKMYQGAGGAGGEDEPPSHD 46
Score = 39.7 bits (91), Expect(3) = 2e-67
Identities = 15/34 (44%), Positives = 24/34 (70%)
Frame = -3
Query: 358 RTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPK 391
R +P + LKDA + +D+D+++LVGG TR P+
Sbjct: 994 RLSSPVEQVLKDAKVKKEDVDDIVLVGGSTRYPQ 893
Score = 21.2 bits (43), Expect(3) = 2e-67
Identities = 8/18 (44%), Positives = 13/18 (71%)
Frame = -2
Query: 346 SKFEALVNNLIERTKAPC 363
+KFE L +L+++T PC
Sbjct: 1031 AKFEELNMDLLKKTLKPC 978
>TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic reticulum
HSC70-cognate binding protein precursor {Glycine max},
partial (38%)
Length = 962
Score = 249 bits (637), Expect = 2e-66
Identities = 134/223 (60%), Positives = 165/223 (73%), Gaps = 8/223 (3%)
Frame = +1
Query: 54 VIGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPE 113
VIGIDLGTT SCV V + + +++ N +G R TPS V+FT E L+G AK A NPE
Sbjct: 295 VIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVSFTDD-ERLIGEAAKNLAAVNPE 471
Query: 114 NTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAKGQQ-----YSPSQIGAF 168
TI KRLIGR+F+D + Q++MK+VPYKIV +G +++ + + +SP +I A
Sbjct: 472 RTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNR-DGKPYIQVRVKDGETKVFSPEEISAM 648
Query: 169 VLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALS 228
+L KMKETAE +LGKT+ AV+TVPAYFNDAQRQATKDAG IAGL V RIINEPTAAA++
Sbjct: 649 ILGKMKETAEGFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAAAIA 828
Query: 229 YGMNKEG---LIAVFDLGGGTFDVSILEISNGVFEVKATNGDT 268
YG++K+G I VFDLGGGTFDVSIL I NGVFEV ATNGDT
Sbjct: 829 YGLDKKGGEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDT 957
>TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock protein
{Arabidopsis thaliana}, partial (92%)
Length = 3027
Score = 224 bits (571), Expect = 8e-59
Identities = 135/404 (33%), Positives = 215/404 (52%), Gaps = 13/404 (3%)
Frame = +1
Query: 54 VIGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPE 113
V+G D G + V+V + VV N E R TP++V F K + +GT + NP+
Sbjct: 244 VVGFDFGNESCIVAVARQRGIDVVLNDESKRETPAIVCFGDK-QRFIGTAGAASTMMNPK 420
Query: 114 NTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAK----GQQYSPSQIGAFV 169
N+IS KRLIG++F DP+ Q+++K +P+ + + P+G + A+ ++++ +Q+ +
Sbjct: 421 NSISQIKRLIGKKFADPELQRDLKSLPFNVTEGPDGYPLIHARYLGESREFTATQVFGMM 600
Query: 170 LTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSY 229
L+ +KE A+ L V I +P YF D QR++ DA IAGL L +I+E TA AL+Y
Sbjct: 601 LSNLKEIAQKNLNAAVVDCCIGIPVYFTDLQRRSVLDAATIAGLHPLHLIHETTATALAY 780
Query: 230 GMNKEGL-------IAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDF 282
G+ K L +A D+G + V I G V + + D LGG DFD AL
Sbjct: 781 GIYKTDLPENEWLNVAFVDVGHASMQVCIAGFKKGQLHVLSHSYDRSLGGRDFDEALFHH 960
Query: 283 LVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNIT 342
+FK ID+ ++ A RLR A EK K LS+ + +N+ + + K +
Sbjct: 961 FAAKFKEEYKIDVYQNARACLRLRAACEKLKKVLSANPEAPLNIECLMDE----KDVRGF 1128
Query: 343 LTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIFGK 402
+ R FE L ++ER K P + L +A ++ ++I V +VG +RVP + ++++ F K
Sbjct: 1129IKRDDFEQLSLPILERVKGPLEKALAEAGLTVENIHMVEVVGSGSRVPAINKILTEFFKK 1308
Query: 403 SPCKGVNPDEAVAMGAALQGGILRG--DVKELLLLDVTPLSLGI 444
P + +N E VA GAALQ IL V+E + + P S+ +
Sbjct: 1309EPRRTMNASECVARGAALQCAILSPTFKVREFQVNESFPFSVSL 1440
>TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [imported] -
Arabidopsis thaliana, partial (91%)
Length = 1717
Score = 143 bits (361), Expect(2) = 4e-54
Identities = 79/231 (34%), Positives = 135/231 (58%), Gaps = 6/231 (2%)
Frame = +1
Query: 305 LREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEALVNNLIERTKAPCQ 364
LR A + A +LS+ S ++++ D + + + R++FE + + E+ ++
Sbjct: 787 LRVATQDAIHQLSTQSSVQVDV-----DLGNSLKICKVVDRAEFEEVNKEVFEKCESLII 951
Query: 365 SCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIFGKSPC-KGVNPDEAVAMGAALQGG 423
CL DA + DI++V+LVGG + +P+V+ +V+ + + KG+NP E GA ++G
Sbjct: 952 QCLHDAKVEVGDINDVILVGGCSYIPRVENLVTNLCKITEVYKGINPLEGAVCGATMEGA 1131
Query: 424 ILRGDVK-----ELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQ 478
+ G +LL + TPL++GI G F +I RNTT+P +K +F+T DNQT+
Sbjct: 1132 VASGISDPFGNLDLLTIQATPLAIGIRADGNKFVPVIPRNTTMPARKDLLFTTIHDNQTE 1311
Query: 479 VGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTV 529
I V +GE + A +N +LG F+++G+P AP+G+P+I V DIDA ++ V
Sbjct: 1312 ALILVYEGEGKKAEENHLLGYFKIMGIPAAPKGVPEISVCMDIDAANVLRV 1464
Score = 87.0 bits (214), Expect(2) = 4e-54
Identities = 74/254 (29%), Positives = 118/254 (46%), Gaps = 24/254 (9%)
Frame = +3
Query: 60 GTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPE---NTI 116
GT+ V+V G ++++N + S V F K E G + + + +TI
Sbjct: 3 GTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTF--KDEAPSGGVTSQFSHEHEMLFGDTI 176
Query: 117 SGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNG-----DAWVEAKGQQYSPSQIGAFVLT 171
KRLIGR DP K +P+ + G A V + +P ++ A L
Sbjct: 177 FNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLAMFLV 353
Query: 172 KMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYGM 231
+++ E +L + + V+TVP F+ Q + A +AGL VLR++ EPTA AL YG
Sbjct: 354 ELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVALLYGQ 533
Query: 232 NK------------EGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNAL 279
+ E + +F++G G DV++ + GV ++KA G T +GGED +
Sbjct: 534 QQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDLLQNM 710
Query: 280 LDFLV----NEFKR 289
+ L+ N FKR
Sbjct: 711 MRHLLPDSENIFKR 752
>TC85397 homologue to GP|170386|gb|AAA99920.1|| glucose-regulated protein 78
{Lycopersicon esculentum}, partial (57%)
Length = 648
Score = 204 bits (519), Expect = 9e-53
Identities = 125/264 (47%), Positives = 155/264 (58%), Gaps = 3/264 (1%)
Frame = +1
Query: 61 TTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPENTISGAK 120
TT SCV V + + +++ N +G R TPS V+FT E L+G AK A NPE TI K
Sbjct: 1 TTYSCVGVYKNGHVEIIANDQGNRITPSWVSFTDD-ERLIGEAAKNLAAVNPERTIFDVK 177
Query: 121 RLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAKGQQYSPSQIGAFVLTKMKETAEAY 180
RLIGR+F+D + Q++MK+VPYKIV N D G+ Y
Sbjct: 178 RLIGRKFEDKEVQRDMKLVPYKIV---NRD------GKPYI------------------- 273
Query: 181 LGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYGMNKEG---LI 237
QATKDAG IAGL V RIINEPTAAA++YG++K+G I
Sbjct: 274 ---------------------QATKDAGVIAGLNVARIINEPTAAAIAYGLDKKGGEKNI 390
Query: 238 AVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRSESIDLSK 297
VFDLGGGTFDVSIL I NGVFEV ATNGDT LGGEDFD + ++ + K+ S D+SK
Sbjct: 391 LVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRIREYFIKLIKKKHSKDISK 570
Query: 298 DKLALQRLREAAEKAKIELSSTSQ 321
D AL +LR+ +E+AK LSS Q
Sbjct: 571 DNRALGKLRKESERAKRALSSQHQ 642
>TC85395 homologue to GP|6911553|emb|CAB72130.1 heat shock protein 70
{Cucumis sativus}, partial (35%)
Length = 723
Score = 196 bits (497), Expect = 3e-50
Identities = 106/222 (47%), Positives = 150/222 (66%), Gaps = 10/222 (4%)
Frame = +3
Query: 430 KELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGERE 489
++LLLLDVTPLSLG+ET GG+ T LI RNTTIPTKK QVFST +DNQ V I+V +GER
Sbjct: 3 QDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERA 182
Query: 490 MASDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITI-KSSG 548
DN +LG+FEL G+PPAPRG+PQI V FDIDANGI+ VSA+DK+TG++ +ITI G
Sbjct: 183 RTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKG 362
Query: 549 GLSDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEKSLSEYR--EKIPAEVAK 606
LS +EI+ MV+EAE + +D+E K +D +N+ + Y++ ++ + + K+ E K
Sbjct: 363 RLSKEEIEKMVEEAEKYKAEDEEHKKKVDAKNALENYAYNMRNTIKDEKIGSKLSPEDKK 542
Query: 607 EIEDAV-------SDLRSAMAGDSADEIKSKLDAANKAVSKI 641
+I+DA+ + A A + D++K N ++K+
Sbjct: 543 KIDDAIDAAIQWLDSNQLAEADEFQDKMKELESLCNPIIAKM 668
>BQ139103 homologue to GP|2655420|gb| heat shock cognate protein HSC70
{Brassica napus}, partial (29%)
Length = 628
Score = 195 bits (496), Expect = 4e-50
Identities = 101/196 (51%), Positives = 134/196 (67%), Gaps = 4/196 (2%)
Frame = +2
Query: 42 SRPFSSRAAGNDVIGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVG 101
++P + + G IGIDLGTT SCV V + +++ N +G RTTPS VAFT E L+G
Sbjct: 47 NKPMAGKGEG-PAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDS-ERLIG 220
Query: 102 TPAKRQAVTNPENTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWV----EAKG 157
AK Q NP NT+ AKRLIGRRF D Q +MK+ P+KI+ P + + +
Sbjct: 221 DAAKNQVAMNPINTVFDAKRLIGRRFSDASVQSDMKLWPFKIISGPAEKPLIGVNYKGED 400
Query: 158 QQYSPSQIGAFVLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLR 217
++++ +I + VL KM+E AEAYLG + AV+TVPAYFND+QRQATKDAG IAGL V+R
Sbjct: 401 KEFAAEEISSMVLMKMREIAEAYLGSAIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMR 580
Query: 218 IINEPTAAALSYGMNK 233
IINEPTAAA++YG++K
Sbjct: 581 IINEPTAAAIAYGLDK 628
>TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chaperone hsc70-3
- tomato, partial (27%)
Length = 667
Score = 174 bits (440), Expect = 1e-43
Identities = 92/182 (50%), Positives = 122/182 (66%), Gaps = 4/182 (2%)
Frame = +3
Query: 55 IGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPEN 114
IGIDLGTT SCV+V + ++ N G RTTPS VAF + E ++G A A +NP N
Sbjct: 108 IGIDLGTTYSCVAVCKNGEIDIIVNDLGKRTTPSFVAF-KDSERMIGDAAFNIAASNPTN 284
Query: 115 TISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEA----KGQQYSPSQIGAFVL 170
TI AKRLIGR+F DP Q ++K+ P+K++ N + + + ++ +I + VL
Sbjct: 285 TIFDAKRLIGRKFSDPIVQSDVKLWPFKVIGDLNDKPMIVVNYNDEEKHFAAEEISSMVL 464
Query: 171 TKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYG 230
KM+E AE +LG V VITVPAYFND+QRQ+T+DAG IAGL V+RIINEPTAAA++YG
Sbjct: 465 VKMREIAETFLGSIVEDVVITVPAYFNDSQRQSTRDAGAIAGLNVMRIINEPTAAAIAYG 644
Query: 231 MN 232
N
Sbjct: 645 FN 650
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.312 0.131 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,525,830
Number of Sequences: 36976
Number of extensions: 158170
Number of successful extensions: 780
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 688
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 694
length of query: 673
length of database: 9,014,727
effective HSP length: 103
effective length of query: 570
effective length of database: 5,206,199
effective search space: 2967533430
effective search space used: 2967533430
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148396.10


