

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148395.6 - phase: 0
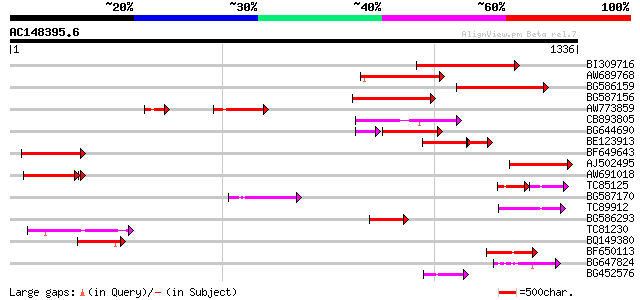
(1336 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 282 7e-76
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 198 1e-50
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 173 5e-43
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 169 5e-42
AW773859 166 4e-41
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 164 2e-40
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 134 8e-38
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 112 3e-35
BF649643 similar to PIR|H86486|H86 protein Ty1/copia-element pol... 135 1e-31
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 120 4e-27
AW691018 weakly similar to GP|2462935|em open reading frame 1 {B... 118 5e-27
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 70 2e-26
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 114 3e-25
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 107 2e-23
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 100 6e-21
TC81230 96 1e-19
BQ149380 94 5e-19
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 92 1e-18
BG647824 weakly similar to PIR|G96722|G96 hypothetical protein F... 85 2e-16
BG452576 weakly similar to GP|12005223|gb|A reverse transcriptas... 69 9e-12
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 282 bits (721), Expect = 7e-76
Identities = 139/242 (57%), Positives = 176/242 (72%)
Frame = +2
Query: 959 QVCKLLKSLYGLKQASRKWYEKLTGFLLLQGYKQSASDHSLFILHSDTCFTALLVYVDDV 1018
+VC+L KS+YGLKQASR+WY KL+ L+ GY QS+SD SLF D+ FT LLVYVDD+
Sbjct: 17 KVCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDDI 196
Query: 1019 ILAGNSMTEIDRIKAVLDVEFKIKDLGKLKYFLGIEVAHSKLGISICQRKYCLDLLKDTG 1078
+LAGN ++EI +K L FKIKDLG L+YFLG+EVA SK GI + QRKY L+LL+D+G
Sbjct: 197 VLAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDSG 376
Query: 1079 LLGAKPVTTPLDPSIKLHQDNSPPHDDILSYRRLVGKLLYLTTTRPDIAFVVQQLSQFLN 1138
L K TP D S+KLH +SP ++D YRRL+GKL+YLTTTRPDI+F VQQLSQF++
Sbjct: 377 NLAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFVS 556
Query: 1139 APTITHYDTACRVVKYLKGTPGQGLLFRRDSQLQLLGFTDADWAGCPDSRRSTSGYCFFL 1198
P HY A RV++YLK P +GL + S L+L F D+DWA CP +R+S +GY FL
Sbjct: 557 KPQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWATCPTTRKSVTGYWVFL 736
Query: 1199 GS 1200
GS
Sbjct: 737 GS 742
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 198 bits (504), Expect = 1e-50
Identities = 97/207 (46%), Positives = 132/207 (62%), Gaps = 9/207 (4%)
Frame = +1
Query: 827 EASQQDC---------WVKAMNSELSALNHNKTWIFVDTPPNIKPIGSKWVYKIKHKSDG 877
E QDC W++AM +E AL NKTW V PP+ K IG KWVY++K DG
Sbjct: 49 EPCPQDCENLPLSDPRWLQAMKTEYKALIDNKTWDLVPLPPHKKAIGCKWVYRVKENPDG 228
Query: 878 TIERYKARLVAKGYTQVEGIDFFDTFSPVAKITTVRILIALASINSWHLHQLDVNNAFLH 937
++ ++KARLVAKG++Q G D+ +TFSPV K T+R+++ +A W + Q+D+NNAFL+
Sbjct: 229 SVNKFKARLVAKGFSQTLGCDYTETFSPVIKPVTIRLILTIAITYKWEIQQIDINNAFLN 408
Query: 938 GDLSENVYMSVPQGVHSPKPNQVCKLLKSLYGLKQASRKWYEKLTGFLLLQGYKQSASDH 997
G L E VYMS PQG + + VCKL KSLYGLKQA R WYE LT + G+ +S D
Sbjct: 409 GFLQEEVYMSQPQGFEAANKSLVCKLNKSLYGLKQAPRAWYEXLTSAQIQFGFTKSRCDP 588
Query: 998 SLFILHSDTCFTALLVYVDDVILAGNS 1024
SL I + + L +YVDD+++ G+S
Sbjct: 589 SLLIYNQNGACIYLXIYVDDILITGSS 669
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 173 bits (438), Expect = 5e-43
Identities = 85/217 (39%), Positives = 133/217 (61%)
Frame = +1
Query: 1053 IEVAHSKLGISICQRKYCLDLLKDTGLLGAKPVTTPLDPSIKLHQDNSPPHDDILSYRRL 1112
+EV ++ GI ICQRKY DLL+ G+ + P+ P KL +D + D Y+++
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 1113 VGKLLYLTTTRPDIAFVVQQLSQFLNAPTITHYDTACRVVKYLKGTPGQGLLFRRDSQLQ 1172
VG L+YL TRPD+ +V+ +S+F+N PT H RV++YL GT G++++R+ +
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEK 360
Query: 1173 LLGFTDADWAGCPDSRRSTSGYCFFLGSSLISWRAKKQHTVARSSSEAEYRALSFASCEL 1232
L +TD+D+AG D R+STSGY F L S +SW +KKQ V S+++AE+ A +F +C+
Sbjct: 361 LEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQS 540
Query: 1233 QWLLYLLKDLRVKCNKLPVLFCDNQSAIHIAGNPVFH 1269
W+ +L+ L + ++CDN S I ++ NPV H
Sbjct: 541 VWMRRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLH 651
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 169 bits (429), Expect = 5e-42
Identities = 84/196 (42%), Positives = 127/196 (63%), Gaps = 1/196 (0%)
Frame = -1
Query: 808 SHQKFALAITDASEPISYKEASQQDCWVKAMNSELSALNHNKTWIFVDTPPNIKPIGSKW 867
+H F + + + P SY+EA + W +++ +E A+ N TW + P K + S+W
Sbjct: 600 AHCAFMVNLNENHIPRSYEEAMEDKEWKESVGAEAGAMIKNDTWYESELPKGKKAVSSRW 421
Query: 868 VYKIKHKSDGTIERYKARLVAKGYTQVEGIDFFDTFSPVAKITTVRILIALASINSWHLH 927
++ IK+K+DG+IER K RLVA+G+T G D+ +TF+PVAK+ T+RI+++LA W L
Sbjct: 420 IFTIKYKADGSIERKKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGLW 241
Query: 928 QLDVNNAFLHGDLSENVYMSVPQGV-HSPKPNQVCKLLKSLYGLKQASRKWYEKLTGFLL 986
Q+DV NAFL G+L + VYM P G+ H K V +L K++YGLKQ+ R WY KL+ L
Sbjct: 240 QMDVKNAFLQGELEDEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTLN 61
Query: 987 LQGYKQSASDHSLFIL 1002
+G+++S DH+LF L
Sbjct: 60 GRGFRKSELDHTLFTL 13
>AW773859
Length = 538
Score = 166 bits (421), Expect = 4e-41
Identities = 83/132 (62%), Positives = 93/132 (69%), Gaps = 2/132 (1%)
Frame = -3
Query: 480 IQDKSNLKMTGLGELIEGLYFLNLGSQVFSQFNHQPVIALSQ--SSSSTTFLPQEALWHF 537
IQ+K +LKM G GEL EGLYFL +Q IA SQ TFLPQEALWHF
Sbjct: 380 IQEKRSLKMIGSGELFEGLYFLT------TQDTPSATIASSQVQPQPQPTFLPQEALWHF 219
Query: 538 RLGHLSNDRLLRMQQHFPCIKVDSNSVCDICHYSRHKKLPFKLSMNKASHCYELIHFDIW 597
RLGHLSN +LL + +FP I +D NSVCDICHYSRHKKLPF+LS N+AS CYEL HFDIW
Sbjct: 218 RLGHLSNRKLLSLHSNFPFITIDQNSVCDICHYSRHKKLPFQLSTNRASKCYELFHFDIW 39
Query: 598 GPISTHSIHGHK 609
GP ST SIH +
Sbjct: 38 GPFSTQSIHNQR 3
Score = 63.9 bits (154), Expect = 4e-10
Identities = 34/59 (57%), Positives = 42/59 (70%)
Frame = -2
Query: 318 NEEREDLDDSKSCKGNSNTEPSFGITKEQYEQLVTLLQSQQASSSKVNLSHVTNHVTSG 376
+E RE+LDDSKSC+G S+G TKEQYE LV LL + S+SKV +SHVT+H SG
Sbjct: 534 HEGREELDDSKSCRGTE----SYGFTKEQYEHLVNLLATPSGSTSKV-VSHVTSHTNSG 373
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 164 bits (416), Expect = 2e-40
Identities = 94/268 (35%), Positives = 144/268 (53%), Gaps = 18/268 (6%)
Frame = +3
Query: 816 ITDASEPISYKEASQQDCWVKAMNSELSALNHNKTWIFVDTPPNIKPIGSKWVYKIKHKS 875
+T S+P +++EA + + W +MN+E+ A N TW D K IG KW++K K
Sbjct: 24 LTMTSDPTTFEEAVKSEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKLNE 203
Query: 876 DGTIERYKARLVAKGYTQVEGIDFFDTFSPVAKITTVRILIALASINSWHLHQLDVNNAF 935
+G IE+YKARLVAKGY+Q G+D+ + F+PVA+ T+R++IALA+ Q+
Sbjct: 204 NGEIEKYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALAA-------QIK----- 347
Query: 936 LHGDLSENVYMSVPQGVHSPKPNQVCKLL-----------------KSLYGLKQASRKWY 978
+ V +S HS N + K L ++LYGLKQA R WY
Sbjct: 348 -----RDGVCIS*M*KAHSCMEN*MRKFLLINHRVM*RRVIS*RVKRALYGLKQAPRAWY 512
Query: 979 EKLTGFLLLQGYKQSASDHSLFI-LHSDTCFTALLVYVDDVILAGNSMTEIDRIKAVLDV 1037
++ + +G+++ +H+LF+ L + +YVDD+I GN + K +
Sbjct: 513 SRIEAYFTKEGFEKCPYEHTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKK 692
Query: 1038 EFKIKDLGKLKYFLGIEVAHSKLGISIC 1065
EF + DLGK+ YFLG+EV ++ GI IC
Sbjct: 693 EFNMSDLGKMHYFLGVEVTQNEKGIYIC 776
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 134 bits (337), Expect(2) = 8e-38
Identities = 67/141 (47%), Positives = 97/141 (68%), Gaps = 1/141 (0%)
Frame = -2
Query: 879 IERYKARLVAKGYTQVEGIDFFDTFSPVAKITTVRILIALASINSWHLHQLDVNNAFLHG 938
I R K++LV +GY Q EGID+ + FSPVA++ +RILIA A+ + L+Q+DV +AF++G
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 939 DLSENVYMSVPQGVHSPK-PNQVCKLLKSLYGLKQASRKWYEKLTGFLLLQGYKQSASDH 997
DL E V++ P G + PN V +L K+LYGLKQA R WYE+L+ FLL G+K+ D+
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 998 SLFILHSDTCFTALLVYVDDV 1018
+LF+L + + VYVDD+
Sbjct: 64 TLFLLKRE*ELLIIQVYVDDI 2
Score = 42.7 bits (99), Expect(2) = 8e-38
Identities = 19/58 (32%), Positives = 29/58 (49%)
Frame = -3
Query: 815 AITDASEPISYKEASQQDCWVKAMNSELSALNHNKTWIFVDTPPNIKPIGSKWVYKIK 872
A + EP + KEA + W+ +M EL +K W V P IG++WV++ K
Sbjct: 606 AFISSIEPKNVKEALRDADWINSMQEELHQFERSKVWYLVPRPEGKTVIGTRWVFRNK 433
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 112 bits (281), Expect(2) = 3e-35
Identities = 60/118 (50%), Positives = 79/118 (66%), Gaps = 1/118 (0%)
Frame = +1
Query: 972 QASRKWYEKLTGFLLLQGYKQSASDHSLFILHSDTCFTALL-VYVDDVILAGNSMTEIDR 1030
Q+ R W+++ T + GY Q +DH++FI HS T A+L VYVDD+ L G+ I R
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKR 180
Query: 1031 IKAVLDVEFKIKDLGKLKYFLGIEVAHSKLGISICQRKYCLDLLKDTGLLGAKPVTTP 1088
+K +L EF+IKDLG LKYFLG+EVA K G SI QRKY LDLLK+T ++G K + P
Sbjct: 181 LKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSISQRKYVLDLLKETRMIGCKTIRDP 354
Score = 55.8 bits (133), Expect(2) = 3e-35
Identities = 29/58 (50%), Positives = 40/58 (68%)
Frame = +2
Query: 1080 LGAKPVTTPLDPSIKLHQDNSPPHDDILSYRRLVGKLLYLTTTRPDIAFVVQQLSQFL 1137
L KP TP+D ++KL ++ D Y+RLVGKL+YL+ TRPDI+FVV +SQF+
Sbjct: 329 LDVKPSETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQFM 502
>BF649643 similar to PIR|H86486|H86 protein Ty1/copia-element polyprotein
[imported] - Arabidopsis thaliana, partial (2%)
Length = 608
Score = 135 bits (339), Expect = 1e-31
Identities = 62/152 (40%), Positives = 94/152 (61%), Gaps = 2/152 (1%)
Frame = +1
Query: 28 NVYYVHASDGPSSVAITPVLSHSNYHSWARSMRRALGAKNKFDFVDGSIPVPTDFDP--N 85
N YY+H SD P + + L+ +NY SW+RSM AL AKNK F++GSI P++ D
Sbjct: 118 NPYYIHPSDHPGHLLVPTKLNGTNYSSWSRSMVHALTAKNKVGFINGSIKTPSEVDQPAE 297
Query: 86 FKAWNRCNMLVHSWIMNSVEDSIAQSIVFLENAIDVWNELKERFSQGDFIRISELQCEIF 145
+ WNRCN ++ SW+ +SVE +A+ ++ + A VW + K++FSQ + I ++Q +
Sbjct: 298 YALWNRCNSMILSWLTHSVEPDLAKGVIHAKTACQVWEDFKDQFSQKNIPAIYQIQKSLA 477
Query: 146 SLKQDSRSVTEFFTALKVLWEELEAYLPTPVC 177
SL Q + S + +FT +K LW+ELE Y P C
Sbjct: 478 SLSQGTVSASTYFTKIKGLWDELETYRTLPTC 573
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 120 bits (301), Expect = 4e-27
Identities = 59/147 (40%), Positives = 94/147 (63%)
Frame = +2
Query: 1179 ADWAGCPDSRRSTSGYCFFLGSSLISWRAKKQHTVARSSSEAEYRALSFASCELQWLLYL 1238
+DWAG ++R+STSGY F LG+ ISW +KKQ VA S++EAEY A + + + WL +
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 1239 LKDLRVKCNKLPVLFCDNQSAIHIAGNPVFHERTKHLEIDCHFVREKLQQNIFKLLPIKS 1298
L+ + + N ++CDN+SAI ++ NPVFH R+KH++I H +RE + + + +
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCPT 361
Query: 1299 QSQLADFFTKPLPPKNFHSFLPKLNMI 1325
+ ++AD FTKPL ++F+ L M+
Sbjct: 362 EEKIADIFTKPLKIESFYKLKKMLGMM 442
>AW691018 weakly similar to GP|2462935|em open reading frame 1 {Brassica
oleracea}, partial (12%)
Length = 639
Score = 118 bits (296), Expect(2) = 5e-27
Identities = 55/133 (41%), Positives = 84/133 (62%)
Frame = +2
Query: 32 VHASDGPSSVAITPVLSHSNYHSWARSMRRALGAKNKFDFVDGSIPVPTDFDPNFKAWNR 91
+H SD P + L+ NY W+RSM +L AKNK +DG++ P+ DP W R
Sbjct: 164 LHHSDTPGISLVNQPLNGGNYGEWSRSMLLSLSAKNKLGLIDGTVKAPSADDPKLPLWKR 343
Query: 92 CNMLVHSWIMNSVEDSIAQSIVFLENAIDVWNELKERFSQGDFIRISELQCEIFSLKQDS 151
CN LV +WI++S+E IA+S++F + A VW++L +RFSQGD RI +++ EI +Q S
Sbjct: 344 CNDLVLTWILHSIEPDIARSVIFSDTAAAVWSDLHDRFSQGDESRIYQIRQEISECRQGS 523
Query: 152 RSVTEFFTALKVL 164
+++++T LK L
Sbjct: 524 LLISDYYTKLKSL 562
Score = 22.3 bits (46), Expect(2) = 5e-27
Identities = 7/18 (38%), Positives = 11/18 (60%)
Frame = +3
Query: 162 KVLWEELEAYLPTPVCAC 179
+V W+EL +Y C+C
Sbjct: 555 RVFWDELGSYQEPITCSC 608
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 70.1 bits (170), Expect(2) = 2e-26
Identities = 37/92 (40%), Positives = 54/92 (58%)
Frame = +3
Query: 1224 ALSFASCELQWLLYLLKDLRVKCNKLPVLFCDNQSAIHIAGNPVFHERTKHLEIDCHFVR 1283
+L A E W+ L+++L K ++ V +CD+QSA+HIA NP FH RTKH+ I HFVR
Sbjct: 228 SLPQACKEAIWMQRLMEELGHKQEQITV-YCDSQSALHIARNPAFHSRTKHIGIQYHFVR 404
Query: 1284 EKLQQNIFKLLPIKSQSQLADFFTKPLPPKNF 1315
E +++ + I + LAD TK + F
Sbjct: 405 EVVEEGSVDMQKIHTNDNLADAMTKSINTDKF 500
Score = 68.9 bits (167), Expect(2) = 2e-26
Identities = 34/75 (45%), Positives = 50/75 (66%)
Frame = +1
Query: 1150 RVVKYLKGTPGQGLLFRRDSQLQLLGFTDADWAGCPDSRRSTSGYCFFLGSSLISWRAKK 1209
R+++Y+KGT G + F S+L + G+ D+D+AG D R+ST+GY F L +SW +K
Sbjct: 7 RIMRYIKGTSGVAVCFG-GSELTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVSWLSKL 183
Query: 1210 QHTVARSSSEAEYRA 1224
Q VA S++EAEY A
Sbjct: 184 QTVVALSTTEAEYMA 228
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 114 bits (284), Expect = 3e-25
Identities = 67/177 (37%), Positives = 99/177 (55%), Gaps = 3/177 (1%)
Frame = -3
Query: 515 PVIALSQSSSSTTFLPQEALWHFRLGHLSNDRLLRMQQHFPCIKVDSNSVCDICHYSRHK 574
PV S +S++ L ++ALWH RLGH + R L + P + V N C+ C +H
Sbjct: 641 PVSNYKCSFTSSSSLNKDALWHARLGH-PHGRALNLM--LPGV-VFENKNCEACILGKHC 474
Query: 575 KLPFKLSMNKASHCYELIHFDIWGPISTHSIHGHKYFITALDDYSRFTWIMLCKTKSEVS 634
K F + +C++LI+ D+W S S HKYF+T +D+ S++TW+ L +K V
Sbjct: 473 KNVFPRTSTVYENCFDLIYTDLWTAPSL-SRDNHKYFVTFIDEKSKYTWLTLIPSKDRVI 297
Query: 635 KSVQNFILNIENQFNCKVKTVRTDNGPEFLMPDFYS---SKGIEHQTSCVETPQQNG 688
+ +NF + N ++ K+K +R+DNG E+ F S GI HQTSC TPQQNG
Sbjct: 296 DAFKNFQAYVTNHYHAKIKILRSDNGGEYTSYAFKSHLDHHGILHQTSCPYTPQQNG 126
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 107 bits (268), Expect = 2e-23
Identities = 59/160 (36%), Positives = 96/160 (59%), Gaps = 2/160 (1%)
Frame = +1
Query: 1151 VVKYLKGTPGQGLLFRRDSQLQ--LLGFTDADWAGCPDSRRSTSGYCFFLGSSLISWRAK 1208
V+KYL + L + + +Q + L G+ DAD+AG D+R+S SG+ F L + ISW+A
Sbjct: 16 VLKYLNESLKSSLKYTKAAQEEDALEGYVDADYAGNVDTRKSLSGFVFTLYGTTISWKAN 195
Query: 1209 KQHTVARSSSEAEYRALSFASCELQWLLYLLKDLRVKCNKLPVLFCDNQSAIHIAGNPVF 1268
+Q V S+++AEY A + WL ++ +L + + + CD+QSAIH+A + V+
Sbjct: 196 QQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGITQEYVKI-HCDSQSAIHLANHQVY 372
Query: 1269 HERTKHLEIDCHFVREKLQQNIFKLLPIKSQSQLADFFTK 1308
HERTKH++I HF+R+ ++ + + S+ AD FTK
Sbjct: 373 HERTKHIDIRLHFIRDMIESKEIVVEKMASEENPADVFTK 492
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 99.8 bits (247), Expect = 6e-21
Identities = 46/90 (51%), Positives = 64/90 (71%)
Frame = +2
Query: 849 KTWIFVDTPPNIKPIGSKWVYKIKHKSDGTIERYKARLVAKGYTQVEGIDFFDTFSPVAK 908
+T V P +KPIG +W+YKIK DGT+ +YKARLVAKGY + +GIDF + F+PV +
Sbjct: 53 QTLKLVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPVVR 232
Query: 909 ITTVRILIALASINSWHLHQLDVNNAFLHG 938
I T+ +L+ALA+ N +H +DV AFL+G
Sbjct: 233 IETI*LLLALAATNGC*IHHIDVKIAFLNG 322
>TC81230
Length = 958
Score = 95.5 bits (236), Expect = 1e-19
Identities = 67/259 (25%), Positives = 109/259 (41%), Gaps = 9/259 (3%)
Frame = +1
Query: 43 ITPVLSHSNYHSWARSMRRALGAKNKFDFVDGSIPVPTD--------FDPNFKAWNRCNM 94
I+ +L+ SNY+ WA SM L + + +V G PT F + W+ N
Sbjct: 226 ISIILNGSNYNHWAESMCGFLKGRRLWRYVTGDKKCPTKGKDDTADAFADKLEEWDSKNH 405
Query: 95 LVHSWIMNSVEDSIAQSIVFLENAIDVWNELKERFSQGDFIRISELQCEIFSLKQDS-RS 153
+ +W N+ SI ENA +VW+ LK+R++ D +L ++ +LKQ S +
Sbjct: 406 QIITWFRNTSIPSIHMQFGRFENAKEVWDHLKQRYTISDLSHQYQLLKDLSNLKQQSGQP 585
Query: 154 VTEFFTALKVLWEELEAYLPTPVCACPHRCMCITGVMNAKHQHEITRSIRFLTGLNDSFD 213
V EF ++V+W +L +C T + + R I+FL L D ++
Sbjct: 586 VYEFLAQMEVIWNQL--------TSCEPSLKDATDMKTYETHRNRVRLIQFLMALTDEYE 741
Query: 214 LVRSQILLMNPLPTINKIFSMVMQHERQFKISIPVEDSTILVNSAGKSQGRGRGNGNSNS 273
VR+ L NPLPT+ + E + ++ P D V +
Sbjct: 742 PVRASSLHQNPLPTLENALPCLKSEETRLQLVPPKADLAFAV----------------TN 873
Query: 274 NGKRSCSFCGRDGHTIDIC 292
N + C C + GH+ C
Sbjct: 874 NATKPCRHCQKSGHSFSDC 930
>BQ149380
Length = 419
Score = 93.6 bits (231), Expect = 5e-19
Identities = 52/117 (44%), Positives = 71/117 (60%), Gaps = 6/117 (5%)
Frame = +3
Query: 161 LKVLWEELEAYLPTPVCACPHRCMCITGVMNAKHQHEITRSIRFLTGLNDSFDLVRSQIL 220
L+ LW+EL+ Y P P C CP +C C + N K R I+FL GLND + V S++L
Sbjct: 72 LRGLWDELDQYRPKPQCTCPIQCTCFA-MRNNKALTAEDRIIKFLMGLNDDYQGVTSKVL 248
Query: 221 LMNPLPTINKIFSMVMQHERQFKISI-----PVEDSTI-LVNSAGKSQGRGRGNGNS 271
LM PLP IN +FSMVMQ ER+ + ++ P++D+T LVN+ + GRG G S
Sbjct: 249 LMYPLPQINMVFSMVMQQERKMQ*NVVSSPTPIDDTTCGLVNAMDGQRQFGRGRGXS 419
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 92.4 bits (228), Expect = 1e-18
Identities = 51/124 (41%), Positives = 75/124 (60%), Gaps = 3/124 (2%)
Frame = +1
Query: 1123 RPDIAFVVQQLSQFLNAPTITHYDTACRVVKYLKGTPGQGLLF---RRDSQLQLLGFTDA 1179
RPDI + V +S+F++ P H A R+++Y++GT GLLF + +L+ ++D+
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAKSEVYELICYSDS 300
Query: 1180 DWAGCPDSRRSTSGYCFFLGSSLISWRAKKQHTVARSSSEAEYRALSFASCELQWLLYLL 1239
DW G RRSTSGY F + ISW KKQ A SS EAEY A +FA+ + WL ++
Sbjct: 301 DWCG---DRRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLDSVI 471
Query: 1240 KDLR 1243
K+L+
Sbjct: 472 KELK 483
>BG647824 weakly similar to PIR|G96722|G96 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (5%)
Length = 721
Score = 84.7 bits (208), Expect = 2e-16
Identities = 62/164 (37%), Positives = 85/164 (51%), Gaps = 6/164 (3%)
Frame = -3
Query: 1141 TITHYDTACRVVKYLKGTPGQGLLFRRDSQLQLLGFTDADWAGCPDSRRSTSGYCFFLGS 1200
T H A +++ YLK +P Q + F S +Q+ F D+D D T
Sbjct: 599 TAQHPQAAIQIL-YLKISPSQ*IFF--PS*IQIKAFCDSD*I---DQAA*T-----LENQ 453
Query: 1201 SLISWRAKKQHTVARSSSEAEYRALSFAS-----CELQWLLYLLKDLRVKCNKLPVLFCD 1255
S+I + H+ A + + Y + S CE++WL YLL DL+ K +L+CD
Sbjct: 452 SVIFASS*ATHSYAGNLKKKRYNFKIYRSI*STICEIKWLTYLLNDLKFTFIKPAMLYCD 273
Query: 1256 NQSAI-HIAGNPVFHERTKHLEIDCHFVREKLQQNIFKLLPIKS 1298
NQSA HIA N F ERTKH+E+DCH VR KLQ +F +L I S
Sbjct: 272 NQSAARHIAANSSFLERTKHIELDCHIVRVKLQLKLFHILHILS 141
>BG452576 weakly similar to GP|12005223|gb|A reverse transcriptase-like protein
{Spiranthes hongkongensis}, partial (25%)
Length = 676
Score = 69.3 bits (168), Expect = 9e-12
Identities = 44/106 (41%), Positives = 64/106 (59%)
Frame = +2
Query: 975 RKWYEKLTGFLLLQGYKQSASDHSLFILHSDTCFTALLVYVDDVILAGNSMTEIDRIKAV 1034
RK Y KL L+ GYKQS +DHS+F FT LVY+DD++ A + +E +K+
Sbjct: 236 RK*Y*KLLSTLISLGYKQSPNDHSIFSFGRR--FTIFLVYLDDIVFARDDHSETQLVKSH 409
Query: 1035 LDVEFKIKDLGKLKYFLGIEVAHSKLGISICQRKYCLDLLKDTGLL 1080
LD F I LG L Y +G+++A S+ I Q KY ++LL+++G L
Sbjct: 410 LDKNFIIIGLGTLHY-VGVKIA*SES*IIDDQCKYTIELLEESGQL 544
Score = 29.3 bits (64), Expect(2) = 0.97
Identities = 40/158 (25%), Positives = 67/158 (42%), Gaps = 14/158 (8%)
Frame = +3
Query: 960 VCKLLKSLYGLKQASRKWYEKLTGFLLLQGYKQSASDHSLFILHSDTCFTALLV------ 1013
VCK S+YGLK A R+ Y K ++ Q + L T F L V
Sbjct: 165 VCKFHNSIYGLKHAHRQ*YSKFFEGNDIESCYQL*FL*DINNLQMITPFLVLAVGLLFFW 344
Query: 1014 YVDDVILAGNSMTEIDRIKAVLDVEFKIKD--LGKLKYFLGIEVAHSKLGISICQRKYCL 1071
Y+ ++ +T I ++ + + +I + G ++ + ++ + + L
Sbjct: 345 YIWMILFL--LVTIILKLNLLSPIWIRIS*S*V*AHSIMSGSKLLNQRVKLLMISANILL 518
Query: 1072 DLLKDT-GLLGAKPVTTPLDPSIKL-----HQDNSPPH 1103
LK LG KP +TP DPS+KL ++ +SP H
Sbjct: 519 SCLKKVVS*LGMKPSSTPRDPSLKLKLYNIYETSSPYH 632
Score = 21.9 bits (45), Expect(2) = 0.97
Identities = 8/11 (72%), Positives = 8/11 (72%)
Frame = +1
Query: 924 WHLHQLDVNNA 934
WH LDVNNA
Sbjct: 40 WHQRLLDVNNA 72
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.134 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 47,848,777
Number of Sequences: 36976
Number of extensions: 781394
Number of successful extensions: 4381
Number of sequences better than 10.0: 89
Number of HSP's better than 10.0 without gapping: 4277
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4352
length of query: 1336
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1228
effective length of database: 5,021,319
effective search space: 6166179732
effective search space used: 6166179732
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC148395.6


