

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148395.10 - phase: 0
(247 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
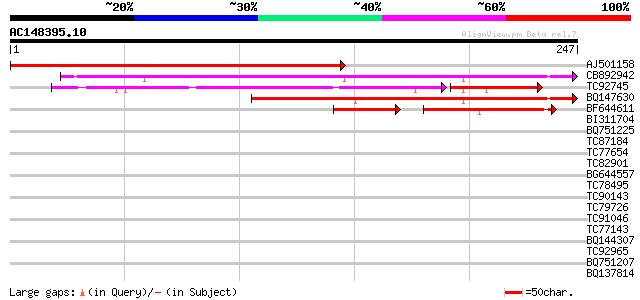
Score E
Sequences producing significant alignments: (bits) Value
AJ501158 similar to GP|9759555|dbj| gb|AAD20416.1~gene_id:MOJ9.1... 299 8e-82
CB892942 similar to GP|13569548|gb unknown {Arabidopsis thaliana... 161 2e-40
TC92745 similar to GP|10177271|dbj|BAB10624. gene_id:MJB21.5~pir... 107 8e-33
BQ147630 similar to GP|9759368|dbj gene_id:MYJ24.9~pir||T00822~s... 119 8e-28
BF644611 similar to GP|11994755|dbj gb|AAB84348.1~gene_id:T5M7.9... 52 5e-12
BI311704 weakly similar to PIR|T04605|T046 hypothetical protein ... 30 0.86
BQ751225 similar to GP|18376350|em probable aldehyde dehydrogena... 30 0.86
TC87184 similar to GP|22296345|dbj|BAC10115. contains EST AU0569... 30 1.1
TC77654 similar to GP|22137236|gb|AAM91463.1 At2g36630/F1O11.26 ... 29 1.5
TC82901 similar to GP|22137236|gb|AAM91463.1 At2g36630/F1O11.26 ... 29 1.5
BG644557 similar to GP|21321777|gb OSJNBa0031O09.02 {Oryza sativ... 28 3.3
TC78495 homologue to PIR|S53504|S53504 extensin-like protein S3 ... 28 4.3
TC90143 weakly similar to GP|9280639|gb|AAF86508.1| F21B7.1 {Ara... 28 4.3
TC79726 similar to GP|17064808|gb|AAL32558.1 Unknown protein {Ar... 28 4.3
TC91046 similar to GP|9279622|dbj|BAB01080.1 gene_id:MOJ10.6~unk... 27 5.6
TC77143 similar to GP|559557|gb|AAA51649.1|| arabinogalactan-pro... 27 5.6
BQ144307 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 27 7.3
TC92965 weakly similar to GP|10764862|gb|AAF24552.2 F1K23.23 {Ar... 27 7.3
BQ751207 weakly similar to GP|604427|gb|AA ACOB protein {Emerice... 27 7.3
BQ137814 weakly similar to PIR|G02127|G021 fus-like protein - hu... 27 9.5
>AJ501158 similar to GP|9759555|dbj| gb|AAD20416.1~gene_id:MOJ9.16~similar to
unknown protein {Arabidopsis thaliana}, partial (22%)
Length = 641
Score = 299 bits (765), Expect = 8e-82
Identities = 146/146 (100%), Positives = 146/146 (100%)
Frame = +3
Query: 1 MAPPHAAQTMTLPTAQRPPPAITSPKVPMQISLQPANSKSKRSSTNKFFGKFRSMFRSFP 60
MAPPHAAQTMTLPTAQRPPPAITSPKVPMQISLQPANSKSKRSSTNKFFGKFRSMFRSFP
Sbjct: 204 MAPPHAAQTMTLPTAQRPPPAITSPKVPMQISLQPANSKSKRSSTNKFFGKFRSMFRSFP 383
Query: 61 IIVPSCKLPTMNGNHRTSETIIHGGTRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAI 120
IIVPSCKLPTMNGNHRTSETIIHGGTRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAI
Sbjct: 384 IIVPSCKLPTMNGNHRTSETIIHGGTRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAI 563
Query: 121 PTGKLLQDMGMGLNRIALECEKHSSN 146
PTGKLLQDMGMGLNRIALECEKHSSN
Sbjct: 564 PTGKLLQDMGMGLNRIALECEKHSSN 641
>CB892942 similar to GP|13569548|gb unknown {Arabidopsis thaliana}, partial
(45%)
Length = 767
Score = 161 bits (408), Expect = 2e-40
Identities = 93/237 (39%), Positives = 133/237 (55%), Gaps = 12/237 (5%)
Frame = +2
Query: 23 TSPKVPMQISLQPANSKSKRSSTNKFFGKFRSMFR--SFPIIVPSCKLPTMNGNHRTSET 80
+SP + S Q + ++T K RS +F I P+C T+ ++
Sbjct: 38 SSPSIS-SFSNQRQQQAKQTTTTKKLSSLIRSFLNIFTFQTIFPTCNWLTIPSTLSSTSI 214
Query: 81 IIHGGTRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALEC 140
G ++TGTLFG R+ ++ A Q + P LLLELA+ T L+++M GL RIALEC
Sbjct: 215 SPSLGRKVTGTLFGNRRGHISFAVQLHPRAEPLLLLELAMSTSSLVKEMSSGLVRIALEC 394
Query: 141 EKHS------SNDKTKIVDEPIWTLFCNGKKMGYGVKRDPTDDDLYVIQMLHAVSVAVGV 194
K S S + ++ EP WT++CNG+K GY V R + D +V+ + +VSV GV
Sbjct: 395 RKTSSTASAVSGGRVRLFHEPDWTMYCNGRKCGYAVSRTCGELDWHVLTTVQSVSVGAGV 574
Query: 195 LP----SDMSDPQDGELSYMRAHFERVIGSKDSETYYMMMPDGNSNGPELSVFFVRV 247
+P +GEL YMRA FERV+GS+DSE +YM+ PDGN GPELS+F +R+
Sbjct: 575 IPMLEDGGGCGGSEGELMYMRAKFERVVGSRDSEAFYMLNPDGN-GGPELSIFLLRI 742
>TC92745 similar to GP|10177271|dbj|BAB10624.
gene_id:MJB21.5~pir||T09377~similar to unknown protein
{Arabidopsis thaliana}, partial (71%)
Length = 810
Score = 107 bits (266), Expect(2) = 8e-33
Identities = 66/179 (36%), Positives = 103/179 (56%), Gaps = 7/179 (3%)
Frame = +1
Query: 19 PPAITSPKVPMQISLQPANSKSKRSST-----NKFF-GKFRSMFRSFPIIVPSCKLPTMN 72
P +SP PM+ PA + R +T NKF G MF+ FP++ CK+ +
Sbjct: 97 PSVHSSPCYPME---NPAIASLLRHTTGDQKRNKFSAGGLLKMFKLFPMLTSGCKMVALL 267
Query: 73 GNHRTSETIIHGGTRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMG 132
G R + + TGT+FGYRK RV++A QED++ P L+EL + T L ++M
Sbjct: 268 GRPRKP---MLKDSATTGTIFGYRKGRVSIAIQEDTRQMPIFLIELPMLTSALNKEMSSD 438
Query: 133 LNRIALECEKHSSNDKTKIVDEPIWTLFCNGKKMGYGVKRDPT-DDDLYVIQMLHAVSV 190
+ RIALE E + +K K+++E +W ++CNG+K+GY ++R D++L V+Q L VS+
Sbjct: 439 IVRIALESE--TKTNKKKLLEEFVWAVYCNGRKVGYSIRRKQMGDEELQVMQHLRGVSM 609
Score = 50.4 bits (119), Expect(2) = 8e-33
Identities = 26/43 (60%), Positives = 32/43 (73%), Gaps = 3/43 (6%)
Frame = +3
Query: 193 GVLP--SDMSDPQDGE-LSYMRAHFERVIGSKDSETYYMMMPD 232
GVLP SD + DG+ ++YMR FERVIGSKDSE +YM+ PD
Sbjct: 618 GVLPTASDHKESSDGDQMTYMRGXFERVIGSKDSEXFYMINPD 746
>BQ147630 similar to GP|9759368|dbj gene_id:MYJ24.9~pir||T00822~similar to
unknown protein {Arabidopsis thaliana}, partial (46%)
Length = 649
Score = 119 bits (299), Expect = 8e-28
Identities = 60/145 (41%), Positives = 92/145 (63%), Gaps = 3/145 (2%)
Frame = +2
Query: 106 EDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECEKHSSNDKT--KIVDEPIWTLFCNG 163
+D L+ELA P L+++M G RIALEC+K +K ++++EP W +CNG
Sbjct: 2 KDPTSQEAFLIELATPISGLVREMASGSVRIALECDKAKEAEKKTLRLLEEPQWRTYCNG 181
Query: 164 KKMGYGVKRDPTDDDLYVIQMLHAVSVAVGVLP-SDMSDPQDGELSYMRAHFERVIGSKD 222
KK G+ +R+ + +++ + +S+ GV+P +D Q+GEL YMRA FER++GS+D
Sbjct: 182 KKCGFANRRECGQKEWDILKAVEPISMGAGVIPGTDNGSEQEGELMYMRAKFERIVGSRD 361
Query: 223 SETYYMMMPDGNSNGPELSVFFVRV 247
SE +YMM PD N PELS++ +RV
Sbjct: 362 SEAFYMMNPDSN-GVPELSIYLLRV 433
>BF644611 similar to GP|11994755|dbj gb|AAB84348.1~gene_id:T5M7.9~similar to
unknown protein {Arabidopsis thaliana}, partial (17%)
Length = 299
Score = 52.4 bits (124), Expect(2) = 5e-12
Identities = 29/61 (47%), Positives = 39/61 (63%), Gaps = 3/61 (4%)
Frame = +3
Query: 181 VIQMLHAVSVAVGVLPSDMSDPQ---DGELSYMRAHFERVIGSKDSETYYMMMPDGNSNG 237
+++ + +S+ GVLP + +GEL YMRA +ERVIGSKDSE +YMM G S G
Sbjct: 120 ILKAVEPISMGAGVLPMPSTGNGGEFEGELMYMRAKYERVIGSKDSEAFYMMNHVG-SGG 296
Query: 238 P 238
P
Sbjct: 297 P 299
Score = 35.0 bits (79), Expect(2) = 5e-12
Identities = 12/29 (41%), Positives = 18/29 (61%)
Frame = +2
Query: 142 KHSSNDKTKIVDEPIWTLFCNGKKMGYGV 170
K K+++EPIW +CNG+K GY +
Sbjct: 2 KGGGRKSLKLLEEPIWRSYCNGRKCGYAL 88
>BI311704 weakly similar to PIR|T04605|T046 hypothetical protein F20O9.30 -
Arabidopsis thaliana, partial (11%)
Length = 504
Score = 30.0 bits (66), Expect = 0.86
Identities = 14/41 (34%), Positives = 21/41 (51%)
Frame = +2
Query: 4 PHAAQTMTLPTAQRPPPAITSPKVPMQISLQPANSKSKRSS 44
P+ +T +PP + + PK+P+ PA S SK SS
Sbjct: 335 PNVTPNVTAVPPSQPPASASPPKIPLTYVDSPAPSPSKASS 457
>BQ751225 similar to GP|18376350|em probable aldehyde dehydrogenase
{Neurospora crassa}, partial (31%)
Length = 568
Score = 30.0 bits (66), Expect = 0.86
Identities = 23/72 (31%), Positives = 33/72 (44%), Gaps = 2/72 (2%)
Frame = -1
Query: 11 TLPTAQRPP--PAITSPKVPMQISLQPANSKSKRSSTNKFFGKFRSMFRSFPIIVPSCKL 68
T PT PP P +P S +PA S S ++T F G+F S+ P++ S
Sbjct: 310 TPPTVWPPPFTPRT*TPPSASATSSRPAPSGS--TATTCFTGRFLSVATRSPVLAESLAR 137
Query: 69 PTMNGNHRTSET 80
P RTS++
Sbjct: 136 PRSQTTPRTSQS 101
>TC87184 similar to GP|22296345|dbj|BAC10115. contains EST
AU056950(S21011)~similar to Arabidopsis thaliana
chromosome 2 full-Length cDNA, partial (58%)
Length = 1309
Score = 29.6 bits (65), Expect = 1.1
Identities = 16/51 (31%), Positives = 24/51 (46%)
Frame = +2
Query: 2 APPHAAQTMTLPTAQRPPPAITSPKVPMQISLQPANSKSKRSSTNKFFGKF 52
APP ++ + P ++ P +T+P P A+ SSTNKF F
Sbjct: 584 APPPSSSAV--PQEKKSIPVVTAPSNPKPKKTDNASKSRSSSSTNKFSSMF 730
>TC77654 similar to GP|22137236|gb|AAM91463.1 At2g36630/F1O11.26
{Arabidopsis thaliana}, partial (94%)
Length = 1939
Score = 29.3 bits (64), Expect = 1.5
Identities = 16/38 (42%), Positives = 21/38 (55%), Gaps = 2/38 (5%)
Frame = +3
Query: 3 PPHAAQTMTLPTAQRPPPAITSPKVPM--QISLQPANS 38
P H++ T + R P TSPK+PM +I L P NS
Sbjct: 75 PKHSSSTSYPVSQSRFSPLFTSPKIPMTTKIHLYPFNS 188
>TC82901 similar to GP|22137236|gb|AAM91463.1 At2g36630/F1O11.26
{Arabidopsis thaliana}, partial (22%)
Length = 647
Score = 29.3 bits (64), Expect = 1.5
Identities = 16/38 (42%), Positives = 21/38 (55%), Gaps = 2/38 (5%)
Frame = +2
Query: 3 PPHAAQTMTLPTAQRPPPAITSPKVPM--QISLQPANS 38
P H++ T + R P TSPK+PM +I L P NS
Sbjct: 77 PKHSSSTSYPVSQSRFSPLFTSPKIPMTTKIHLYPFNS 190
>BG644557 similar to GP|21321777|gb OSJNBa0031O09.02 {Oryza sativa (japonica
cultivar-group)}, partial (22%)
Length = 693
Score = 28.1 bits (61), Expect = 3.3
Identities = 18/67 (26%), Positives = 29/67 (42%)
Frame = -2
Query: 158 TLFCNGKKMGYGVKRDPTDDDLYVIQMLHAVSVAVGVLPSDMSDPQDGELSYMRAHFERV 217
T K+G G++ D +L V +H VA L + +PQ L YMR ++
Sbjct: 458 TYIQGSSKLGIGIRNDARRSNLRVDSSVHYEIVAAAYLGDVVKEPQ--WLQYMRQWGPKI 285
Query: 218 IGSKDSE 224
+ +E
Sbjct: 284 VYDSKTE 264
>TC78495 homologue to PIR|S53504|S53504 extensin-like protein S3 - alfalfa,
partial (97%)
Length = 1049
Score = 27.7 bits (60), Expect = 4.3
Identities = 15/36 (41%), Positives = 17/36 (46%)
Frame = +3
Query: 3 PPHAAQTMTLPTAQRPPPAITSPKVPMQISLQPANS 38
PP Q P PPP +SP P+Q S PA S
Sbjct: 297 PPTTPQASPPPVQSSPPPVQSSPP-PLQSSPPPAQS 401
>TC90143 weakly similar to GP|9280639|gb|AAF86508.1| F21B7.1 {Arabidopsis
thaliana}, partial (5%)
Length = 724
Score = 27.7 bits (60), Expect = 4.3
Identities = 12/29 (41%), Positives = 19/29 (65%)
Frame = +2
Query: 97 KARVNLAFQEDSKCHPFLLLELAIPTGKL 125
+A + AFQ ++ C P+LLL + + T KL
Sbjct: 77 RAGMREAFQRENICGPYLLLLMGLKTSKL 163
>TC79726 similar to GP|17064808|gb|AAL32558.1 Unknown protein {Arabidopsis
thaliana}, partial (36%)
Length = 777
Score = 27.7 bits (60), Expect = 4.3
Identities = 21/56 (37%), Positives = 26/56 (45%), Gaps = 10/56 (17%)
Frame = -1
Query: 19 PPAITSPKVPMQISLQPANSKSKRSSTNKF----------FGKFRSMFRSFPIIVP 64
PPAI P QISL P S + STN+F G+ F+ F +IVP
Sbjct: 582 PPAIDEK--PDQISLFPLPSTNLERSTNRFPLGHTQE*FVVGEIPK*FQRFSLIVP 421
>TC91046 similar to GP|9279622|dbj|BAB01080.1 gene_id:MOJ10.6~unknown
protein {Arabidopsis thaliana}, partial (6%)
Length = 978
Score = 27.3 bits (59), Expect = 5.6
Identities = 14/58 (24%), Positives = 28/58 (48%)
Frame = -3
Query: 26 KVPMQISLQPANSKSKRSSTNKFFGKFRSMFRSFPIIVPSCKLPTMNGNHRTSETIIH 83
K+P + LQP K + ++ + R++ + P+ PS LP G + + ++H
Sbjct: 418 KIPWRQLLQPLEKKEQMQASGQTMKSQRALQQQLPL--PSYYLPQKFGKEKMTWKVVH 251
>TC77143 similar to GP|559557|gb|AAA51649.1|| arabinogalactan-protein {Pyrus
communis}, partial (30%)
Length = 956
Score = 27.3 bits (59), Expect = 5.6
Identities = 16/44 (36%), Positives = 21/44 (47%), Gaps = 1/44 (2%)
Frame = +3
Query: 3 PPHAAQ-TMTLPTAQRPPPAITSPKVPMQISLQPANSKSKRSST 45
PP A T T PTA PP A +P P + PA + + + T
Sbjct: 249 PPTATPPTATPPTATPPPAAAPTPATPAPATSPPAPTPTSDAPT 380
>BQ144307 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (24%)
Length = 1059
Score = 26.9 bits (58), Expect = 7.3
Identities = 10/26 (38%), Positives = 13/26 (49%)
Frame = -2
Query: 3 PPHAAQTMTLPTAQRPPPAITSPKVP 28
PPH P + PPP + SP +P
Sbjct: 479 PPHPLPISPPPPLRPPPPPLPSPDIP 402
>TC92965 weakly similar to GP|10764862|gb|AAF24552.2 F1K23.23 {Arabidopsis
thaliana}, partial (25%)
Length = 697
Score = 26.9 bits (58), Expect = 7.3
Identities = 16/50 (32%), Positives = 24/50 (48%), Gaps = 1/50 (2%)
Frame = +2
Query: 4 PHAAQTMTLPTAQRPPPAITSPKVPMQISLQPANSKSK-RSSTNKFFGKF 52
PH++ T T P P TSP P S P + + + +S +N F +F
Sbjct: 71 PHSSPTATPPNPFSSPHPSTSPTPPPPSSSSPVHHQRQFQSPSNNFCKQF 220
>BQ751207 weakly similar to GP|604427|gb|AA ACOB protein {Emericella
nidulans}, partial (12%)
Length = 758
Score = 26.9 bits (58), Expect = 7.3
Identities = 17/39 (43%), Positives = 18/39 (45%)
Frame = +1
Query: 7 AQTMTLPTAQRPPPAITSPKVPMQISLQPANSKSKRSST 45
A T T+ PPPAITSP ANS S S T
Sbjct: 250 AHTPTISPPPPPPPAITSPSSTPPRPSSSANSPS*LSLT 366
>BQ137814 weakly similar to PIR|G02127|G021 fus-like protein - human
(fragment), partial (8%)
Length = 1050
Score = 26.6 bits (57), Expect = 9.5
Identities = 15/43 (34%), Positives = 21/43 (47%), Gaps = 2/43 (4%)
Frame = +2
Query: 2 APPHAAQTMTLPTAQRPPPAITSPK--VPMQISLQPANSKSKR 42
A PH AQ T R PP +P+ +P + S + N +S R
Sbjct: 248 ASPHRAQPWPPATPSRAPPHFHTPQHNIPSESSARRKNRRSVR 376
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,762,086
Number of Sequences: 36976
Number of extensions: 109428
Number of successful extensions: 756
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 718
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 741
length of query: 247
length of database: 9,014,727
effective HSP length: 94
effective length of query: 153
effective length of database: 5,538,983
effective search space: 847464399
effective search space used: 847464399
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC148395.10


