

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148360.2 + phase: 0 /pseudo
(322 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
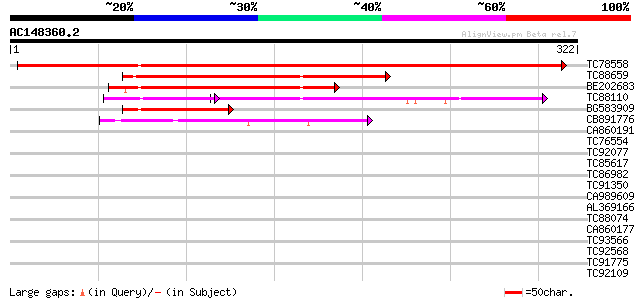
Score E
Sequences producing significant alignments: (bits) Value
TC78558 similar to PIR|T09914|T09914 protein-arginine N-methyltr... 627 e-180
TC88659 similar to GP|16930707|gb|AAL32019.1 At1g04870/F13M7_12 ... 150 6e-37
BE202683 homologue to GP|9293956|dbj protein arginine N-methyltr... 137 6e-33
TC88110 similar to GP|10177192|dbj|BAB10326. arginine methyltran... 106 4e-32
BG583909 similar to GP|12322039|gb| arginine N-methyltransferase... 66 2e-11
CB891776 similar to PIR|G71432|G71 hypothetical protein dl4310w ... 57 7e-09
CA860191 homologue to GP|19697342|gb hypothetical protein {Dicty... 39 0.004
TC76554 similar to GP|7295688|gb|AAF50994.1| CG11931 gene produc... 38 0.005
TC92077 homologue to GP|23497650|gb|AAN37190.1 hypothetical prot... 37 0.008
TC85617 similar to GP|2369766|emb|CAA04664.1 hypothetical protei... 35 0.039
TC86982 similar to GP|10946337|gb|AAG24863.1 CONSTANS-like prote... 35 0.039
TC91350 similar to GP|22831129|dbj|BAC15990. putative protein ar... 35 0.051
CA989609 GP|21959900|gb| hypothetical {Yersinia pestis KIM}, par... 33 0.15
AL369166 similar to GP|3341693|gb|A unknown protein {Arabidopsis... 33 0.19
TC88074 similar to PIR|T00442|T00442 probable rRNA (adenine-N6 N... 33 0.19
CA860177 similar to GP|20976627|gb CG5792 PROTEIN. 6/101 {Dictyo... 33 0.19
TC93566 similar to GP|13272425|gb|AAK17151.1 unknown protein {Ar... 32 0.25
TC92568 weakly similar to SP|Q9DB76|CGX2_MOUSE Protein CGI-112 h... 32 0.33
TC91775 homologue to GP|23495145|gb|AAN35476.1 hypothetical prot... 32 0.43
TC92109 similar to GP|19683031|gb|AAL92653.1 HYPOTHETICAL 31.4 K... 32 0.43
>TC78558 similar to PIR|T09914|T09914 protein-arginine N-methyltransferase
(EC 2.1.1.23) - Arabidopsis thaliana, partial (83%)
Length = 1348
Score = 627 bits (1617), Expect = e-180
Identities = 309/312 (99%), Positives = 310/312 (99%)
Frame = +2
Query: 5 KNNNQTNNTNMDNNNNNNNNHLRFEDADEVIEDVAETSNLDQSMGGCDLDDSNDKTSADY 64
KNNNQTNNTNMDNNNNNNNNHLRFEDADEVIEDVAETSNLDQSMGGCDLDDSNDKTSADY
Sbjct: 2 KNNNQTNNTNMDNNNNNNNNHLRFEDADEVIEDVAETSNLDQSMGGCDLDDSNDKTSADY 181
Query: 65 YFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGA 124
YFDSYSHFG H+EMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGA
Sbjct: 182 YFDSYSHFG-IHEEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGA 358
Query: 125 AHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIIISEWMGYFLLFE 184
AHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIIISEWMGYFLLFE
Sbjct: 359 AHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIIISEWMGYFLLFE 538
Query: 185 NMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWNNVYGFDMSCIKKQA 244
NMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWNNVYGFDMSCIKKQA
Sbjct: 539 NMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWNNVYGFDMSCIKKQA 718
Query: 245 LMEPLVDTVDQNQIATNCQLLKSMDISKMSSGDCSFTAPFKLVAARDDFIHAFVAYFDVS 304
LMEPLVDTVDQNQIATNCQLLKSMDISKMSSGDCSFTAPFKLVAARDDFIHAFVAYFDVS
Sbjct: 719 LMEPLVDTVDQNQIATNCQLLKSMDISKMSSGDCSFTAPFKLVAARDDFIHAFVAYFDVS 898
Query: 305 FTKCHKLMGFST 316
FTKCHKLMGFST
Sbjct: 899 FTKCHKLMGFST 934
>TC88659 similar to GP|16930707|gb|AAL32019.1 At1g04870/F13M7_12
{Arabidopsis thaliana}, partial (41%)
Length = 622
Score = 150 bits (379), Expect = 6e-37
Identities = 76/152 (50%), Positives = 110/152 (72%)
Frame = +1
Query: 65 YFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGA 124
YF +YS F KEML D VR Y N +++N+ FK+KVVLDVGAG+GIL+++ A+AGA
Sbjct: 157 YFCTYS-FLYHQKEMLSDRVRMDAYFNAVFENKHHFKDKVVLDVGAGSGILAIWSAQAGA 333
Query: 125 AHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIIISEWMGYFLLFE 184
VYAVE + M++ A+ +V++N +V+ V++G +E++ LP KVD+IISEWMGYFLL E
Sbjct: 334 KKVYAVEATKMSEHARALVKSNNLEQVVEVIEGSVEDVTLP-EKVDVIISEWMGYFLLRE 510
Query: 185 NMLNSVLFARDKWLVDDGVILPDIASLYLTAI 216
+M +SV+ ARD+WL GV+ P A +++ I
Sbjct: 511 SMFDSVICARDRWLKPTGVMYPSHARMWMAPI 606
>BE202683 homologue to GP|9293956|dbj protein arginine
N-methyltransferase-like protein {Arabidopsis thaliana},
partial (32%)
Length = 592
Score = 137 bits (345), Expect = 6e-33
Identities = 70/134 (52%), Positives = 96/134 (71%), Gaps = 3/134 (2%)
Frame = +2
Query: 57 NDKTSADY---YFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTG 113
N DY YF SY+H G H+EM+KD VRT+TY+ I +++ KVV+DVG GTG
Sbjct: 194 NRSPCTDYDMAYFHSYAHLG-IHQEMIKDRVRTETYREAIMRHQSFIAGKVVVDVGCGTG 370
Query: 114 ILSLFCAKAGAAHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIII 173
ILS+FCA+AGA VYAV+ S +A +A E+V+ N S V+ VL G++E++E+ +VD+II
Sbjct: 371 ILSIFCAQAGAKRVYAVDASDIALQANEVVKANNLSDVVIVLHGRVEDVEID-EEVDVII 547
Query: 174 SEWMGYFLLFENML 187
SEWMGY LL+E+ML
Sbjct: 548 SEWMGYMLLYESML 589
>TC88110 similar to GP|10177192|dbj|BAB10326. arginine
methyltransferase-like protein {Arabidopsis thaliana},
partial (64%)
Length = 1792
Score = 106 bits (264), Expect(2) = 4e-32
Identities = 69/202 (34%), Positives = 112/202 (55%), Gaps = 11/202 (5%)
Frame = +1
Query: 115 LSLFCAKAGAAHVYAVECSHMADRAKEIVETNGY-SKVITVLKGKIEELELPVPKVDIII 173
LSLF A+AGA HVYA+E S MA+ A++++ N + ITV+KG++E++ELP K DI+I
Sbjct: 586 LSLFAAQAGAKHVYAIEASEMAEYARKLIAGNPLLGQRITVIKGRVEDVELP-EKADILI 762
Query: 174 SEWMGYFLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKED---KIEFW- 229
SE MG L+ E ML S + ARD++L +G + P +++ D+ + K FW
Sbjct: 763 SEPMGTLLVNEKMLESYVIARDRFLTPNGKMFPTSGRIHMAPFSDEYLFVEIASKGLFWQ 942
Query: 230 -NNVYGFDMSCIKKQALM----EPLVDTVDQNQIATNCQLLKSMDISKMSSGDC-SFTAP 283
N +G D++ + K A +P+VD D ++ + + MD +K+ + P
Sbjct: 943 QQNYFGVDLTPLHKTAFQGYFSQPVVDAFDP-RLLISPPMCHPMDFTKIKEEELYEIDIP 1119
Query: 284 FKLVAARDDFIHAFVAYFDVSF 305
+ +A+ +H +FDV F
Sbjct: 1120LRFIASVGARVHGLACWFDVLF 1185
Score = 49.3 bits (116), Expect(2) = 4e-32
Identities = 26/66 (39%), Positives = 39/66 (58%)
Frame = +3
Query: 54 DDSNDKTSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTG 113
D+ + +SA YF Y + + ML+D VRT +Y + +NR F +VV+DVGAG+G
Sbjct: 405 DEKIEPSSAKMYFHYYGQLLH-QQNMLQDYVRTGSYYAAVMENRTDFFGRVVVDVGAGSG 581
Query: 114 ILSLFC 119
I + C
Sbjct: 582 IFVIIC 599
>BG583909 similar to GP|12322039|gb| arginine N-methyltransferase 3
putative; 35335-37803 {Arabidopsis thaliana}, partial
(11%)
Length = 707
Score = 66.2 bits (160), Expect = 2e-11
Identities = 35/63 (55%), Positives = 40/63 (62%)
Frame = +1
Query: 65 YFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGA 124
YF SYS FG H+EML D R Y I +N L VV+DVG GTGILSLF A+AGA
Sbjct: 415 YFGSYSSFG-IHREMLSDKARMDAYGQAILKNPSLLNGAVVMDVGCGTGILSLFSAQAGA 591
Query: 125 AHV 127
+ V
Sbjct: 592 SRV 600
>CB891776 similar to PIR|G71432|G71 hypothetical protein dl4310w -
Arabidopsis thaliana, partial (27%)
Length = 531
Score = 57.4 bits (137), Expect = 7e-09
Identities = 43/161 (26%), Positives = 79/161 (48%), Gaps = 6/161 (3%)
Frame = +1
Query: 52 DLDDSNDKTSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAG 111
D DD +++T + F + +ML D+ R ++ I + + + VLD+GAG
Sbjct: 61 DEDDDDEQT---FQFHQPLLATTSYLDMLNDSTRNTAFREAI--EKTITEPCHVLDIGAG 225
Query: 112 TGILSLFCAKAGAAHVYAVECSH---MADRAKEIVETNGYSKVITVLKGKIEELELPVPK 168
TG+LS+ ++A C M K+++ NG I V+ + +ELE+ V
Sbjct: 226 TGLLSMMASRAMGGKGTVTACESYLPMVKLMKKVMRINGLEGRIKVINKRSDELEVGVDL 405
Query: 169 ---VDIIISEWMGYFLLFENMLNSVLFARDKWLVDDGVILP 206
D+++SE + LL E ++ ++ A D LV++ + +P
Sbjct: 406 SSCADVLVSEILDSELLGEGLIPTLQHAHDNLLVENPLTVP 528
>CA860191 homologue to GP|19697342|gb hypothetical protein {Dictyostelium
discoideum}, partial (1%)
Length = 504
Score = 38.5 bits (88), Expect = 0.004
Identities = 17/40 (42%), Positives = 25/40 (62%)
Frame = +2
Query: 6 NNNQTNNTNMDNNNNNNNNHLRFEDADEVIEDVAETSNLD 45
+NN NN+N +NNNN +N+H +F D ++ E NLD
Sbjct: 308 SNNNNNNSNNNNNNNEDNHHNKFNSEDAMLPQTQE-FNLD 424
>TC76554 similar to GP|7295688|gb|AAF50994.1| CG11931 gene product
{Drosophila melanogaster}, partial (42%)
Length = 861
Score = 38.1 bits (87), Expect = 0.005
Identities = 15/27 (55%), Positives = 19/27 (69%)
Frame = -2
Query: 2 GSRKNNNQTNNTNMDNNNNNNNNHLRF 28
GS N++ NN N +NNNNNNNN+ F
Sbjct: 566 GSSSNSSTNNNNNNNNNNNNNNNNTHF 486
Score = 34.3 bits (77), Expect = 0.066
Identities = 13/24 (54%), Positives = 18/24 (74%)
Frame = -2
Query: 2 GSRKNNNQTNNTNMDNNNNNNNNH 25
GS ++N + N N +NNNNNNNN+
Sbjct: 572 GSGSSSNSSTNNNNNNNNNNNNNN 501
Score = 29.6 bits (65), Expect = 1.6
Identities = 12/23 (52%), Positives = 14/23 (60%)
Frame = -2
Query: 3 SRKNNNQTNNTNMDNNNNNNNNH 25
S NNN NN N +NNNN + H
Sbjct: 548 STNNNNNNNNNNNNNNNNTHFIH 480
Score = 28.5 bits (62), Expect = 3.6
Identities = 10/23 (43%), Positives = 17/23 (73%)
Frame = -2
Query: 3 SRKNNNQTNNTNMDNNNNNNNNH 25
S + ++N++ +NNNNNNNN+
Sbjct: 581 SNGGSGSSSNSSTNNNNNNNNNN 513
>TC92077 homologue to GP|23497650|gb|AAN37190.1 hypothetical protein
{Plasmodium falciparum 3D7}, partial (1%)
Length = 376
Score = 37.4 bits (85), Expect = 0.008
Identities = 14/20 (70%), Positives = 16/20 (80%)
Frame = +1
Query: 6 NNNQTNNTNMDNNNNNNNNH 25
NNN +N N +NNNNNNNNH
Sbjct: 4 NNNDNDNNNDNNNNNNNNNH 63
Score = 35.0 bits (79), Expect = 0.039
Identities = 13/21 (61%), Positives = 16/21 (75%)
Frame = +1
Query: 6 NNNQTNNTNMDNNNNNNNNHL 26
N+N NN N ++NNNNNN HL
Sbjct: 28 NDNNNNNNNNNHNNNNNNTHL 90
Score = 35.0 bits (79), Expect = 0.039
Identities = 13/20 (65%), Positives = 16/20 (80%)
Frame = +1
Query: 6 NNNQTNNTNMDNNNNNNNNH 25
NNN NN N +NN+NNNNN+
Sbjct: 22 NNNDNNNNNNNNNHNNNNNN 81
Score = 34.7 bits (78), Expect = 0.051
Identities = 13/20 (65%), Positives = 16/20 (80%)
Frame = +1
Query: 6 NNNQTNNTNMDNNNNNNNNH 25
+NN N+ N DNNNNNNNN+
Sbjct: 1 DNNNDNDNNNDNNNNNNNNN 60
Score = 32.7 bits (73), Expect = 0.19
Identities = 12/20 (60%), Positives = 16/20 (80%)
Frame = +1
Query: 6 NNNQTNNTNMDNNNNNNNNH 25
N+N NN N +NNNNNN+N+
Sbjct: 10 NDNDNNNDNNNNNNNNNHNN 69
Score = 32.7 bits (73), Expect = 0.19
Identities = 12/20 (60%), Positives = 16/20 (80%)
Frame = +1
Query: 6 NNNQTNNTNMDNNNNNNNNH 25
+NN NN N +NNN+NNNN+
Sbjct: 19 DNNNDNNNNNNNNNHNNNNN 78
Score = 31.2 bits (69), Expect = 0.56
Identities = 11/20 (55%), Positives = 16/20 (80%)
Frame = +1
Query: 6 NNNQTNNTNMDNNNNNNNNH 25
N+N +N N +NNNN+NNN+
Sbjct: 16 NDNNNDNNNNNNNNNHNNNN 75
Score = 30.8 bits (68), Expect = 0.73
Identities = 11/20 (55%), Positives = 16/20 (80%)
Frame = +1
Query: 6 NNNQTNNTNMDNNNNNNNNH 25
NN+ NN + +NNNNNNN++
Sbjct: 7 NNDNDNNNDNNNNNNNNNHN 66
Score = 30.4 bits (67), Expect = 0.96
Identities = 11/24 (45%), Positives = 16/24 (65%)
Frame = +1
Query: 6 NNNQTNNTNMDNNNNNNNNHLRFE 29
NNN NN N +NNNNN + + ++
Sbjct: 34 NNNNNNNNNHNNNNNNTHLFITYK 105
Score = 30.0 bits (66), Expect = 1.3
Identities = 11/22 (50%), Positives = 14/22 (63%)
Frame = +1
Query: 3 SRKNNNQTNNTNMDNNNNNNNN 24
+ +NN NN N NNNNNN +
Sbjct: 22 NNNDNNNNNNNNNHNNNNNNTH 87
Score = 29.6 bits (65), Expect = 1.6
Identities = 10/23 (43%), Positives = 17/23 (73%)
Frame = +1
Query: 3 SRKNNNQTNNTNMDNNNNNNNNH 25
+ +N+ N+ N +NNNNN+NN+
Sbjct: 4 NNNDNDNNNDNNNNNNNNNHNNN 72
>TC85617 similar to GP|2369766|emb|CAA04664.1 hypothetical protein {Citrus x
paradisi}, partial (85%)
Length = 1489
Score = 35.0 bits (79), Expect = 0.039
Identities = 20/62 (32%), Positives = 29/62 (46%)
Frame = +1
Query: 2 GSRKNNNQTNNTNMDNNNNNNNNHLRFEDADEVIEDVAETSNLDQSMGGCDLDDSNDKTS 61
G+ + N +N N +NNNNNN N+ D AET ++ +GG +ND
Sbjct: 436 GNVQLNKDPHNNNSNNNNNNNENNSNATDKRFKTLPAAETLPRNEVLGGYIFVCNNDTMQ 615
Query: 62 AD 63
D
Sbjct: 616 ED 621
>TC86982 similar to GP|10946337|gb|AAG24863.1 CONSTANS-like protein {Ipomoea
nil}, partial (54%)
Length = 1643
Score = 35.0 bits (79), Expect = 0.039
Identities = 28/94 (29%), Positives = 44/94 (46%), Gaps = 7/94 (7%)
Frame = +1
Query: 6 NNNQTNNTNMDNNNNNNNNHLRFEDADEVIEDVAETSNLDQSMGGCDLDDSNDKTSADYY 65
NNN NN + D+N NN +L + DE + D+ + + S GG D++ T+ ++
Sbjct: 580 NNNSNNNISNDHNQVANNGYLFSGEVDEYL-DLVDCN----SWGG---DENTFTTNNTHH 735
Query: 66 FDSYS-------HFGNFHKEMLKDTVRTKTYQNV 92
D YS H+G K + D+V Q V
Sbjct: 736 HDEYSQQQQQQDHYGVPQKSYVGDSVVPVQQQQV 837
>TC91350 similar to GP|22831129|dbj|BAC15990. putative protein arginine
N-methyltransferase {Oryza sativa (japonica
cultivar-group)}, partial (13%)
Length = 749
Score = 34.7 bits (78), Expect = 0.051
Identities = 16/31 (51%), Positives = 21/31 (67%)
Frame = +3
Query: 81 KDTVRTKTYQNVIYQNRFLFKNKVVLDVGAG 111
+D VRT TY + +NR F +VV+DVGAG
Sbjct: 657 RDYVRTGTYYAAVIENRADFTGRVVVDVGAG 749
>CA989609 GP|21959900|gb| hypothetical {Yersinia pestis KIM}, partial (18%)
Length = 708
Score = 33.1 bits (74), Expect = 0.15
Identities = 13/21 (61%), Positives = 16/21 (75%)
Frame = +1
Query: 5 KNNNQTNNTNMDNNNNNNNNH 25
+N N NN N +NNNNNNNN+
Sbjct: 445 QNINDYNNNNNNNNNNNNNNN 507
Score = 32.3 bits (72), Expect = 0.25
Identities = 12/19 (63%), Positives = 15/19 (78%)
Frame = +1
Query: 7 NNQTNNTNMDNNNNNNNNH 25
N+ NN N +NNNNNNNN+
Sbjct: 454 NDYNNNNNNNNNNNNNNNN 510
Score = 31.6 bits (70), Expect = 0.43
Identities = 12/19 (63%), Positives = 14/19 (73%)
Frame = +1
Query: 6 NNNQTNNTNMDNNNNNNNN 24
N+ NN N +NNNNNNNN
Sbjct: 454 NDYNNNNNNNNNNNNNNNN 510
>AL369166 similar to GP|3341693|gb|A unknown protein {Arabidopsis thaliana},
partial (13%)
Length = 505
Score = 32.7 bits (73), Expect = 0.19
Identities = 13/25 (52%), Positives = 17/25 (68%)
Frame = +2
Query: 2 GSRKNNNQTNNTNMDNNNNNNNNHL 26
G+ NNN N N + NNNNNNN++
Sbjct: 206 GNTINNNDNNIINNNGNNNNNNNNI 280
Score = 27.3 bits (59), Expect = 8.1
Identities = 12/22 (54%), Positives = 13/22 (58%)
Frame = +2
Query: 4 RKNNNQTNNTNMDNNNNNNNNH 25
RK N NN N NNN NNN+
Sbjct: 200 RKGNTINNNDNNIINNNGNNNN 265
>TC88074 similar to PIR|T00442|T00442 probable rRNA (adenine-N6
N6-)-dimethyltransferase - Arabidopsis thaliana, partial
(83%)
Length = 1394
Score = 32.7 bits (73), Expect = 0.19
Identities = 28/97 (28%), Positives = 46/97 (46%), Gaps = 5/97 (5%)
Frame = +2
Query: 74 NFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGAAHVYAVECS 133
+FHK + ++ + I Q + VVL++G GTG L+ +AG V AVE
Sbjct: 146 SFHKSKGQHILKNPLLVDTIVQKSGIKTTDVVLEIGPGTGNLTKKLLEAG-KKVIAVEID 322
Query: 134 HMADRAKEIVETNGY-----SKVITVLKGKIEELELP 165
+ ++E N S +TV++G + + ELP
Sbjct: 323 -----PRMVLELNKRFQGTPSSRLTVIQGDVLKTELP 418
>CA860177 similar to GP|20976627|gb CG5792 PROTEIN. 6/101 {Dictyostelium
discoideum}, partial (2%)
Length = 360
Score = 32.7 bits (73), Expect = 0.19
Identities = 12/19 (63%), Positives = 15/19 (78%)
Frame = +2
Query: 6 NNNQTNNTNMDNNNNNNNN 24
NN NN N++NNNNNNN+
Sbjct: 35 NNQNQNNKNINNNNNNNNS 91
Score = 30.4 bits (67), Expect = 0.96
Identities = 12/19 (63%), Positives = 13/19 (68%)
Frame = +2
Query: 6 NNNQTNNTNMDNNNNNNNN 24
+NNQ N NNNNNNNN
Sbjct: 32 SNNQNQNNKNINNNNNNNN 88
>TC93566 similar to GP|13272425|gb|AAK17151.1 unknown protein {Arabidopsis
thaliana}, partial (17%)
Length = 640
Score = 32.3 bits (72), Expect = 0.25
Identities = 12/23 (52%), Positives = 16/23 (69%)
Frame = +3
Query: 6 NNNQTNNTNMDNNNNNNNNHLRF 28
+NN N N+ NNN NNNN++ F
Sbjct: 411 HNNNKNKNNIINNNTNNNNNMSF 479
>TC92568 weakly similar to SP|Q9DB76|CGX2_MOUSE Protein CGI-112 homolog.
[Mouse] {Mus musculus}, partial (24%)
Length = 577
Score = 32.0 bits (71), Expect = 0.33
Identities = 13/34 (38%), Positives = 19/34 (55%)
Frame = +3
Query: 1 MGSRKNNNQTNNTNMDNNNNNNNNHLRFEDADEV 34
+G NN NN N DN+N N+NN +D ++
Sbjct: 66 LGDTSINNNKNNKNSDNSNENDNNDDNNDDNGQI 167
>TC91775 homologue to GP|23495145|gb|AAN35476.1 hypothetical protein
{Plasmodium falciparum 3D7}, partial (2%)
Length = 757
Score = 31.6 bits (70), Expect = 0.43
Identities = 14/25 (56%), Positives = 17/25 (68%)
Frame = +1
Query: 7 NNQTNNTNMDNNNNNNNNHLRFEDA 31
NN NN N +NNNNNNN L + +A
Sbjct: 346 NNVINNNNNNNNNNNNNKLL*YINA 420
Score = 28.1 bits (61), Expect = 4.8
Identities = 11/18 (61%), Positives = 13/18 (72%)
Frame = +1
Query: 5 KNNNQTNNTNMDNNNNNN 22
+NN NN N +NNNNNN
Sbjct: 343 RNNVINNNNNNNNNNNNN 396
Score = 27.7 bits (60), Expect = 6.2
Identities = 11/21 (52%), Positives = 14/21 (66%)
Frame = +1
Query: 5 KNNNQTNNTNMDNNNNNNNNH 25
K + N N +NNNNNNNN+
Sbjct: 331 KYTTRNNVINNNNNNNNNNNN 393
>TC92109 similar to GP|19683031|gb|AAL92653.1 HYPOTHETICAL 31.4 KDA PROTEIN
{Dictyostelium discoideum}, partial (2%)
Length = 653
Score = 31.6 bits (70), Expect = 0.43
Identities = 13/24 (54%), Positives = 16/24 (66%)
Frame = +3
Query: 4 RKNNNQTNNTNMDNNNNNNNNHLR 27
R N N +NN +NN +NNN HLR
Sbjct: 360 RDNINYSNNNYNNNNKSNNNKHLR 431
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.135 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,896,484
Number of Sequences: 36976
Number of extensions: 130763
Number of successful extensions: 2215
Number of sequences better than 10.0: 95
Number of HSP's better than 10.0 without gapping: 1158
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1674
length of query: 322
length of database: 9,014,727
effective HSP length: 96
effective length of query: 226
effective length of database: 5,465,031
effective search space: 1235097006
effective search space used: 1235097006
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148360.2


