

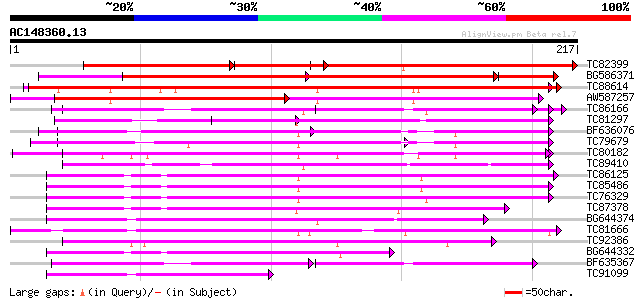
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148360.13 + phase: 0
(217 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC82399 similar to GP|7527717|gb|AAF63166.1| T5E21.6 {Arabidopsi... 176 5e-90
BG586371 similar to PIR|T04273|T04 hypothetical protein F20B18.2... 277 6e-83
TC88614 similar to GP|20161078|dbj|BAB90009. putative mitochondr... 194 2e-50
AW587257 similar to PIR|T04273|T04 hypothetical protein F20B18.2... 149 6e-37
TC86166 similar to PIR|T01729|T01729 mitochondrial solute carrie... 102 9e-23
TC81297 similar to GP|9758252|dbj|BAB08751.1 calcium-binding tra... 91 3e-19
BF636076 similar to GP|3068714|gb| unknown {Arabidopsis thaliana... 87 6e-18
TC79679 similar to GP|18377839|gb|AAL67106.1 AT5g01500/F7A7_20 {... 86 1e-17
TC80182 similar to GP|10177187|dbj|BAB10321. mitochondrial carri... 86 1e-17
TC89410 similar to GP|20260666|gb|AAM13231.1 unknown protein {Ar... 82 2e-16
TC86125 similar to PIR|T04608|T04608 ADP ATP carrier protein F20... 80 5e-16
TC85486 homologue to GP|2780194|emb|CAA05979.1 adenine nucleotid... 78 3e-15
TC76329 homologue to GP|2780194|emb|CAA05979.1 adenine nucleotid... 75 2e-14
TC87378 similar to PIR|S21313|S21313 ADP ATP carrier protein - A... 70 5e-13
BG644374 similar to GP|21536790|gb ADP/ATP translocase-like prot... 69 1e-12
TC81666 similar to PIR|T07793|T07793 uncoupling protein (clone S... 65 2e-11
TC92386 similar to GP|10177187|dbj|BAB10321. mitochondrial carri... 64 4e-11
BG644332 SP|P27081|ADT2 ADP ATP carrier protein mitochondrial p... 63 1e-10
BF635367 similar to PIR|T01729|T01 mitochondrial solute carrier ... 62 1e-10
TC91099 similar to GP|10177844|dbj|BAB11273. ADP/ATP translocase... 59 1e-09
>TC82399 similar to GP|7527717|gb|AAF63166.1| T5E21.6 {Arabidopsis
thaliana}, partial (44%)
Length = 833
Score = 176 bits (445), Expect(3) = 5e-90
Identities = 97/127 (76%), Positives = 99/127 (77%), Gaps = 25/127 (19%)
Frame = +2
Query: 116 KKSIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQV-------------------------Q 150
+K+ AKLTCGSVAGLLGQTFTY LEVVRRQMQV Q
Sbjct: 371 RKASWAKLTCGSVAGLLGQTFTYPLEVVRRQMQVTCFIWSAGNYRWILSNMIQS*PV*VQ 550
Query: 151 NLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLRV 210
NLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLRV
Sbjct: 551 NLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLRV 730
Query: 211 PSRDEVD 217
PSRDEVD
Sbjct: 731 PSRDEVD 751
Score = 117 bits (293), Expect(3) = 5e-90
Identities = 58/58 (100%), Positives = 58/58 (100%)
Frame = +3
Query: 29 AKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIKGIYRGV 86
AKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIKGIYRGV
Sbjct: 3 AKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIKGIYRGV 176
Score = 76.6 bits (187), Expect(3) = 5e-90
Identities = 34/36 (94%), Positives = 35/36 (96%)
Frame = +1
Query: 87 APTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAK 122
APTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIM +
Sbjct: 283 APTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMGE 390
>BG586371 similar to PIR|T04273|T04 hypothetical protein F20B18.290 -
Arabidopsis thaliana, partial (71%)
Length = 861
Score = 277 bits (709), Expect(2) = 6e-83
Identities = 137/144 (95%), Positives = 141/144 (97%)
Frame = +3
Query: 44 QVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYF 103
++VSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGI+G+YRGVAPTLFGIFPYAGLKFYF
Sbjct: 351 KIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAPTLFGIFPYAGLKFYF 530
Query: 104 YEEMKRHVPEDYKKSIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGT 163
YEEMKR VPEDYKKSIMAKLTCGSVAGLLGQTFTY LEVVRRQMQVQNL ASEEAELKGT
Sbjct: 531 YEEMKRRVPEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELKGT 710
Query: 164 MRSMVLIAQKQGWKTLFSGLSINY 187
MRSMVLIAQKQGWKTLFSGLSINY
Sbjct: 711 MRSMVLIAQKQGWKTLFSGLSINY 782
Score = 89.0 bits (219), Expect = 1e-18
Identities = 52/104 (50%), Positives = 62/104 (59%)
Frame = +2
Query: 12 IPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLS 71
IPLFAKELLAGGLAGGFAKTVVAPLERLKILFQ E G+ +
Sbjct: 59 IPLFAKELLAGGLAGGFAKTVVAPLERLKILFQT-----------RRTEFRSAGLSGSVR 205
Query: 72 KTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDY 115
+ K G+ G YRG ++ I PYAGL F YEE +R + + +
Sbjct: 206 RIAKTEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAF 337
Score = 47.4 bits (111), Expect(2) = 6e-83
Identities = 22/23 (95%), Positives = 23/23 (99%)
Frame = +2
Query: 188 IKVVPSAAIGFTVYDTMKSYLRV 210
+KVVPSAAIGFTVYDTMKSYLRV
Sbjct: 782 LKVVPSAAIGFTVYDTMKSYLRV 850
>TC88614 similar to GP|20161078|dbj|BAB90009. putative mitochondrial carrier
{Oryza sativa (japonica cultivar-group)}, partial (74%)
Length = 1412
Score = 194 bits (493), Expect = 2e-50
Identities = 105/217 (48%), Positives = 147/217 (67%), Gaps = 13/217 (5%)
Frame = +1
Query: 8 ILDHIPLFAK----ELLAGGLAGGFAKTVVAPLE--RLKILFQVVSPTKLNVSGM--VNN 59
IL++ P+ +LLAG AGG + PL+ R K+ +QVV G+ V++
Sbjct: 619 ILNNYPMLGTGPSIDLLAGSAAGGTSVLCTYPLDLARTKLAYQVVDTKGCIKDGIKAVHS 798
Query: 60 EQV---YRGIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYK 116
+ V + GI+ L YKE G++G+YRGV PTL GI PYAGLKFY YE++K HVPE+++
Sbjct: 799 QPVGPVHNGIKGVLKSAYKEAGVRGLYRGVGPTLTGILPYAGLKFYTYEKLKMHVPEEHQ 978
Query: 117 KSIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLP--ASEEAELKGTMRSMVLIAQKQ 174
KSI+ +L+CG++AGL GQT TY L+VV++QMQV +L A+ +A K T + I + Q
Sbjct: 979 KSILMRLSCGALAGLFGQTLTYPLDVVKKQMQVGSLQNGANGDAAYKNTFDGLRKIVRNQ 1158
Query: 175 GWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLRVP 211
GW+ LF+G+SINYI++VPSAAI FT YD MK++L VP
Sbjct: 1159GWRQLFAGVSINYIRIVPSAAISFTTYDMMKAWLGVP 1269
Score = 85.5 bits (210), Expect = 1e-17
Identities = 64/223 (28%), Positives = 99/223 (43%), Gaps = 20/223 (8%)
Frame = +1
Query: 6 ESILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRG 65
ES D +P++ KEL+AGG AG AKT VAPLER+KIL+Q G G
Sbjct: 337 ESSFDRVPVYVKELIAGGFAGALAKTSVAPLERVKILWQT------RTGGFHT-----LG 483
Query: 66 IRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYK---KSIMAK 122
+ ++K K G G+Y+G ++ I PYA L F YE K + +Y
Sbjct: 484 VCQSVNKLLKHEGFLGLYKGNGASVIRIVPYAALHFMTYERYKSWILNNYPMLGTGPSID 663
Query: 123 LTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPAS-----------------EEAELKGTMR 165
L GS AG TY L++ R ++ Q + +KG ++
Sbjct: 664 LLAGSAAGGTSVLCTYPLDLARTKLAYQVVDTKGCIKDGIKAVHSQPVGPVHNGIKGVLK 843
Query: 166 SMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 208
S ++ G + L+ G+ ++P A + F Y+ +K ++
Sbjct: 844 SAY---KEAGVRGLYRGVGPTLTGILPYAGLKFYTYEKLKMHV 963
>AW587257 similar to PIR|T04273|T04 hypothetical protein F20B18.290 -
Arabidopsis thaliana, partial (55%)
Length = 630
Score = 149 bits (377), Expect = 6e-37
Identities = 72/92 (78%), Positives = 81/92 (87%), Gaps = 2/92 (2%)
Frame = +3
Query: 18 ELLAGGLAGGFAKTVVAPLE--RLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYK 75
+L+AG L+GG A PL+ R K+ +Q+VSPTKLNVSGMVNNEQVYRGIRDCLSKTYK
Sbjct: 354 DLMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYK 533
Query: 76 EGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEM 107
EGGI+G+YRGVAPTLFGIFPYAGLKFYFYEEM
Sbjct: 534 EGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEM 629
Score = 126 bits (317), Expect = 6e-30
Identities = 84/216 (38%), Positives = 114/216 (51%), Gaps = 12/216 (5%)
Frame = +3
Query: 1 MAASSESILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNE 60
MAASSESILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQ E
Sbjct: 15 MAASSESILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQT-----------RRTE 161
Query: 61 QVYRGIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYK---K 117
G+ + + K G+ G YRG ++ I PYAGL F YEE +R + + + K
Sbjct: 162 FRSAGLSGSVRRIAKTEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWK 341
Query: 118 SIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLP---------ASEEAELKGTMRSMV 168
L GS++G FTY L+++R ++ Q + + E +G +
Sbjct: 342 GPTLDLMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLS 521
Query: 169 LIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTM 204
++ G + L+ G++ + P A + F Y+ M
Sbjct: 522 KTYKEGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEM 629
>TC86166 similar to PIR|T01729|T01729 mitochondrial solute carrier protein
homolog - Arabidopsis thaliana, partial (98%)
Length = 1789
Score = 102 bits (255), Expect = 9e-23
Identities = 58/201 (28%), Positives = 107/201 (52%), Gaps = 9/201 (4%)
Frame = +3
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 76
K L+AGG+AGG ++T VAPLER+KIL QV +P + SG + L ++
Sbjct: 456 KSLVAGGVAGGVSRTAVAPLERMKILLQVQNPHNIKYSGTIQG----------LKYIWRT 605
Query: 77 GGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHV---------PEDYKKSIMAKLTCGS 127
G +G+++G I P + +KF+ YE+ + + ED + + + +L G+
Sbjct: 606 EGFRGLFKGNGTNCARIVPNSAVKFFSYEQASKGILHLYRQQTGNEDAQLTPVLRLGAGA 785
Query: 128 VAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINY 187
AG++ + TY +++VR ++ VQ + +G ++ + +++G + L+ G +
Sbjct: 786 CAGIIAMSATYPMDMVRGRITVQT--EKSPYQYRGMFHALSTVLREEGPRALYKGWLPSV 959
Query: 188 IKVVPSAAIGFTVYDTMKSYL 208
I V+P + F VY+++K YL
Sbjct: 960 IGVIPYVGLNFAVYESLKDYL 1022
Score = 97.4 bits (241), Expect = 4e-21
Identities = 62/212 (29%), Positives = 101/212 (47%), Gaps = 19/212 (8%)
Frame = +3
Query: 21 AGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIK 80
AG AG A + P++ ++ V + + YRG+ LS +E G +
Sbjct: 777 AGACAGIIAMSATYPMDMVRGRITVQTE---------KSPYQYRGMFHALSTVLREEGPR 929
Query: 81 GIYRGVAPTLFGIFPYAGLKFYFYEEMKRHV--------PEDYKKSIMAKLTCGSVAGLL 132
+Y+G P++ G+ PY GL F YE +K ++ +D + S+ +L CG+ AG +
Sbjct: 930 ALYKGWLPSVIGVIPYVGLNFAVYESLKDYLIKTKPFGLAQDSELSVTTRLACGAAAGTI 1109
Query: 133 GQTFTYFLEVVRRQMQVQNLPASEEA-----------ELKGTMRSMVLIAQKQGWKTLFS 181
GQT Y L+V+RR+MQ+ + E G + + + +G+ L+
Sbjct: 1110GQTVAYPLDVIRRRMQMTGWHNAASVITGDGVGKVPLEYTGMVDAFRKTVRHEGFGALYK 1289
Query: 182 GLSINYIKVVPSAAIGFTVYDTMKSYLRVPSR 213
GL N +KVVPS A+ F Y+ +K L V R
Sbjct: 1290GLVPNSVKVVPSIALAFVSYEMVKDILGVEIR 1385
Score = 45.4 bits (106), Expect = 2e-05
Identities = 27/85 (31%), Positives = 46/85 (53%)
Frame = +3
Query: 118 SIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWK 177
SI L G VAG + +T LE ++ +QVQN + GT++ + I + +G++
Sbjct: 447 SICKSLVAGGVAGGVSRTAVAPLERMKILLQVQN---PHNIKYSGTIQGLKYIWRTEGFR 617
Query: 178 TLFSGLSINYIKVVPSAAIGFTVYD 202
LF G N ++VP++A+ F Y+
Sbjct: 618 GLFKGNGTNCARIVPNSAVKFFSYE 692
>TC81297 similar to GP|9758252|dbj|BAB08751.1 calcium-binding
transporter-like protein {Arabidopsis thaliana}, partial
(26%)
Length = 636
Score = 91.3 bits (225), Expect = 3e-19
Identities = 47/135 (34%), Positives = 80/135 (58%), Gaps = 4/135 (2%)
Frame = +1
Query: 78 GIKGIYRGVAPTLFGIFPYAGLKFYFYEEMK----RHVPEDYKKSIMAKLTCGSVAGLLG 133
G + YRG+ P++ G+ PYAG+ FY+ +K +++ D + +L CG+++G LG
Sbjct: 37 GPRAFYRGLLPSVIGMIPYAGIDLAFYDTLKDMSKKYIIHDSDPGPLVQLGCGTISGTLG 216
Query: 134 QTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPS 193
T Y L+V+R ++Q Q L +S+ KG + Q +G++ + GL N +KVVP+
Sbjct: 217 ATCVYPLQVIRTRLQAQPLNSSDA--YKGMFDAFCRTFQHEGFRGFYKGLLPNLLKVVPA 390
Query: 194 AAIGFTVYDTMKSYL 208
A+I + VY++MK L
Sbjct: 391 ASITYMVYESMKKNL 435
Score = 52.0 bits (123), Expect = 2e-07
Identities = 25/94 (26%), Positives = 47/94 (49%)
Frame = +1
Query: 18 ELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEG 77
+L G ++G T V PL+ ++ Q +N+ Y+G+ D +T++
Sbjct: 181 QLGCGTISGTLGATCVYPLQVIRTRLQAQP---------LNSSDAYKGMFDAFCRTFQHE 333
Query: 78 GIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHV 111
G +G Y+G+ P L + P A + + YE MK+++
Sbjct: 334 GFRGFYKGLLPNLLKVVPAASITYMVYESMKKNL 435
>BF636076 similar to GP|3068714|gb| unknown {Arabidopsis thaliana}, partial
(55%)
Length = 673
Score = 86.7 bits (213), Expect = 6e-18
Identities = 59/199 (29%), Positives = 104/199 (51%), Gaps = 2/199 (1%)
Frame = +3
Query: 12 IPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLS 71
+P A LAG +AG AKTV APL+R+K+L Q V + + G + +S
Sbjct: 84 VPKDAALFLAGAVAGAAAKTVTAPLDRIKLLMQT-----HGVRIRQESAKKTIGFIEAIS 248
Query: 72 KTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR-HVPEDYKKSIMAKLTCGSVAG 130
KE GI+G ++G P + + PY+ ++ + YE K+ ++ + S++ +L G+ AG
Sbjct: 249 MIGKEEGIRGYWKGNLPQVIRVIPYSAVQLFAYEIYKKIFKGKNDELSVVGRLAAGAFAG 428
Query: 131 LLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVL-IAQKQGWKTLFSGLSINYIK 189
+ TY L+V+R ++ V+ P TM + L + +++G+ + + GL + I
Sbjct: 429 MTSTFVTYPLDVLRLRLAVE--PGCR------TMTEVALSMVREEGFASFYYGLGPSLIG 584
Query: 190 VVPSAAIGFTVYDTMKSYL 208
+ P A+ F V+D +K L
Sbjct: 585 IAPYIAVNFCVFDLLKKSL 641
Score = 44.7 bits (104), Expect = 3e-05
Identities = 28/99 (28%), Positives = 44/99 (44%)
Frame = +3
Query: 19 LLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGG 78
L AG AG + V PL+ L++ V E R + + +E G
Sbjct: 405 LAAGAFAGMTSTFVTYPLDVLRLRLAV--------------EPGCRTMTEVALSMVREEG 542
Query: 79 IKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKK 117
Y G+ P+L GI PY + F ++ +K+ +PE Y+K
Sbjct: 543 FASFYYGLGPSLIGIAPYIAVNFCVFDLLKKSLPEKYQK 659
>TC79679 similar to GP|18377839|gb|AAL67106.1 AT5g01500/F7A7_20 {Arabidopsis
thaliana}, partial (61%)
Length = 1003
Score = 85.5 bits (210), Expect = 1e-17
Identities = 59/204 (28%), Positives = 104/204 (50%), Gaps = 4/204 (1%)
Frame = +3
Query: 9 LDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIR- 67
L +P A AG AG AKT APL+R+K+L Q V E + I
Sbjct: 285 LAFVPRDAALFAAGAFAGAAAKTFTAPLDRIKLLMQTHGV-------RVGQESAKKAISF 443
Query: 68 -DCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR-HVPEDYKKSIMAKLTC 125
+ ++ KE GI+G ++G P + + PY+ ++ + YE K+ ++ + S++A+L+
Sbjct: 444 VEAITVIGKEEGIRGYWKGNLPQVIRVIPYSAVQLFAYELYKKLFTGQNGELSVVARLSA 623
Query: 126 GSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVL-IAQKQGWKTLFSGLS 184
G+ AG+ TY L+V+R ++ V+ P TM + L + +++G+ + + GL
Sbjct: 624 GAFAGMTSTFITYPLDVLRLRLAVE--PGYR------TMSEVALCMLREEGFASFYKGLG 779
Query: 185 INYIKVVPSAAIGFTVYDTMKSYL 208
+ I + P A+ F V+D +K L
Sbjct: 780 PSLIAIAPYIAVNFCVFDLLKKSL 851
Score = 60.5 bits (145), Expect = 5e-10
Identities = 37/135 (27%), Positives = 61/135 (44%)
Frame = +3
Query: 19 LLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGG 78
L AG AG + + PL+ L++ V E YR + + +E G
Sbjct: 615 LSAGAFAGMTSTFITYPLDVLRLRLAV--------------EPGYRTMSEVALCMLREEG 752
Query: 79 IKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLTCGSVAGLLGQTFTY 138
Y+G+ P+L I PY + F ++ +K+ +PE Y+K + ++ L Y
Sbjct: 753 FASFYKGLGPSLIAIAPYIAVNFCVFDLLKKSLPEKYQKRTETSILTAVLSASLATLTCY 932
Query: 139 FLEVVRRQMQVQNLP 153
L+ VRRQMQ++ P
Sbjct: 933 PLDTVRRQMQLRGTP 977
>TC80182 similar to GP|10177187|dbj|BAB10321. mitochondrial carrier
protein-like {Arabidopsis thaliana}, partial (88%)
Length = 1293
Score = 85.5 bits (210), Expect = 1e-17
Identities = 57/197 (28%), Positives = 99/197 (49%), Gaps = 10/197 (5%)
Frame = +1
Query: 21 AGGLAGGFAKTVVAPLERLKILFQV-VSPTKL--NVSGMVNNEQVYRGIRDCLSKTYKEG 77
AG ++G ++TV +PL+ +KI FQV + PT L ++ ++ Y G+ ++E
Sbjct: 109 AGAISGAISRTVTSPLDVIKIRFQVQLEPTSLWTSLHRDLSKPSKYTGMFQATKDIFREE 288
Query: 78 GIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLT------CGSVAGL 131
GI+G +RG P L + PY ++F ++K K L+ G+VAG
Sbjct: 289 GIRGFWRGNVPALLMVMPYTAIQFTVLHKLKTFASGSSKTENHINLSPYLSYVSGAVAGC 468
Query: 132 LGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVL-IAQKQGWKTLFSGLSINYIKV 190
+Y +++R + Q E ++ MRS + I Q +G++ L+SGLS +++
Sbjct: 469 AATVGSYPFDLLRTILASQG-----EPKVYPNMRSASIDILQTRGFRGLYSGLSPTLVEI 633
Query: 191 VPSAAIGFTVYDTMKSY 207
+P A + F YDT K +
Sbjct: 634 IPYAGLQFGTYDTFKRW 684
Score = 76.3 bits (186), Expect = 9e-15
Identities = 57/226 (25%), Positives = 93/226 (40%), Gaps = 19/226 (8%)
Frame = +1
Query: 2 AASSESILDHIPLFAKELLAGGLAGGFAKTVVA-PLERLKILFQVVSPTKLNVSGMVNNE 60
A+ S +HI L G G A TV + P + L+ + K
Sbjct: 388 ASGSSKTENHINLSPYLSYVSGAVAGCAATVGSYPFDLLRTILASQGEPK---------- 537
Query: 61 QVYRGIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR----------- 109
VY +R + G +G+Y G++PTL I PYAGL+F Y+ KR
Sbjct: 538 -VYPNMRSASIDILQTRGFRGLYSGLSPTLVEIIPYAGLQFGTYDTFKRWTMAWNQVQYS 714
Query: 110 HVPEDYKKSIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPAS-------EEAELKG 162
+ + S CG AG + + L+VV+++ Q++ L E +
Sbjct: 715 NTAAEESISSFQLFLCGLAAGTCAKLVCHPLDVVKKRFQIEGLQRHPRYGARVEHRAYRN 894
Query: 163 TMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 208
++ I QK+GW L+ G+ + +K P+ A+ F Y+ +L
Sbjct: 895 MFDALKRILQKEGWAGLYKGIVPSTVKAAPAGAVTFVAYELTSDWL 1032
>TC89410 similar to GP|20260666|gb|AAM13231.1 unknown protein {Arabidopsis
thaliana}, partial (66%)
Length = 1183
Score = 81.6 bits (200), Expect = 2e-16
Identities = 58/191 (30%), Positives = 97/191 (50%), Gaps = 3/191 (1%)
Frame = +2
Query: 21 AGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIK 80
+G AG A T V PL+ +K FQV P N G V + ++ K G++
Sbjct: 335 SGASAGVIAATFVCPLDVIKTRFQVGVPQLAN--GTVKGSVIVASLQQIFHKE----GLR 496
Query: 81 GIYRGVAPTLFGIFPYAGLKFYFYEEMKRHV---PEDYKKSIMAKLTCGSVAGLLGQTFT 137
G+YRG+APT+ + P + F YE++K + E + S+ + + + AG FT
Sbjct: 497 GMYRGLAPTVLALLPNWAVYFTMYEQLKSLLHSDDESHHLSVGSNMVAAAGAGAATTLFT 676
Query: 138 YFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIG 197
L VV+ ++Q Q + S + T+ ++ IA ++G + L+SGL + + + AI
Sbjct: 677 NPLWVVKTRLQTQGM-RSGVVPYRSTLSALRRIASEEGIRGLYSGL-VPALAGISHVAIQ 850
Query: 198 FTVYDTMKSYL 208
F +Y+T+K YL
Sbjct: 851 FPMYETIKFYL 883
Score = 35.8 bits (81), Expect = 0.013
Identities = 21/71 (29%), Positives = 38/71 (52%)
Frame = +2
Query: 20 LAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGI 79
+A ++ FA T+ P E ++ ++L G ++E+ Y G+ DC+ K +++ G+
Sbjct: 935 IASSVSKIFASTLTYPHEVVR--------SRLQEQGH-HSEKRYSGMIDCIRKVFQQEGV 1087
Query: 80 KGIYRGVAPTL 90
G YRG A L
Sbjct: 1088PGFYRGCATNL 1120
>TC86125 similar to PIR|T04608|T04608 ADP ATP carrier protein F20O9.60 -
Arabidopsis thaliana, partial (81%)
Length = 1640
Score = 80.5 bits (197), Expect = 5e-16
Identities = 55/203 (27%), Positives = 95/203 (46%), Gaps = 7/203 (3%)
Frame = +3
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTY 74
F + L GG++ +KT AP+ER+K+L Q + ++ SG ++ + Y+GI DC ++T
Sbjct: 426 FLIDFLMGGVSAAVSKTAAAPIERVKLLIQ--NQDEMLKSGRLS--EPYKGIGDCFARTM 593
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR-----HVPEDYKKSIMAKLTCGSVA 129
K+ G+ ++RG + FP L F F + KR + Y K L G A
Sbjct: 594 KDEGVIALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDRDGYWKWFAGNLASGGAA 773
Query: 130 GLLGQTFTYFLEVVRRQMQVQNLPASE--EAELKGTMRSMVLIAQKQGWKTLFSGLSINY 187
G F Y L+ R ++ A + E + G + Q G L+ G +I+
Sbjct: 774 GASSLLFVYSLDYARTRLANDAKAAKKGGERQFNGMVDVYKKTLQSDGIAGLYRGFNISC 953
Query: 188 IKVVPSAAIGFTVYDTMKSYLRV 210
+ ++ + F +YD++K + V
Sbjct: 954 VGIIVYRGLYFGMYDSLKPVVLV 1022
>TC85486 homologue to GP|2780194|emb|CAA05979.1 adenine nucleotide
translocator {Lupinus albus}, complete
Length = 1611
Score = 77.8 bits (190), Expect = 3e-15
Identities = 53/201 (26%), Positives = 92/201 (45%), Gaps = 7/201 (3%)
Frame = +1
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTY 74
F + L GG++ +KT AP+ER+K+L Q + ++ SG ++ + Y+GI DC +T
Sbjct: 346 FVIDFLMGGVSAAVSKTAAAPIERIKLLIQ--NQDEMIKSGRLS--EPYKGIGDCFKRTM 513
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR-----HVPEDYKKSIMAKLTCGSVA 129
+ G+ ++RG + FP L F F + KR + Y K L G A
Sbjct: 514 ADEGVVALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDRDGYWKWFAGNLASGGAA 693
Query: 130 GLLGQTFTYFLEVVRRQMQVQNLPASE--EAELKGTMRSMVLIAQKQGWKTLFSGLSINY 187
G F Y L+ R ++ + A + E + G + G L+ G +I+
Sbjct: 694 GASSLLFVYSLDYARTRLANDSKAAKKGGERQFNGLVDVYKKTLATDGVAGLYRGFNISC 873
Query: 188 IKVVPSAAIGFTVYDTMKSYL 208
+ ++ + F +YD++K L
Sbjct: 874 VGIIVYRGLYFGLYDSLKPVL 936
>TC76329 homologue to GP|2780194|emb|CAA05979.1 adenine nucleotide
translocator {Lupinus albus}, partial (81%)
Length = 1497
Score = 75.1 bits (183), Expect = 2e-14
Identities = 53/202 (26%), Positives = 91/202 (44%), Gaps = 8/202 (3%)
Frame = +1
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTY 74
F + L GG++ +KT AP+ER+K+L Q + ++ +G ++ + Y+GI DC +T
Sbjct: 283 FLVDFLMGGVSAAVSKTAAAPIERIKLLIQ--NQDEMIKAGRLS--EPYKGIGDCFKRTT 450
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR-----HVPEDYKKSIMAKLTCGSVA 129
E G+ ++RG + FP L F F + KR + Y K L G A
Sbjct: 451 AEEGVVALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDKDGYWKWFAGNLASGGAA 630
Query: 130 GLLGQTFTYFLEVVRRQMQVQNLPASEEA---ELKGTMRSMVLIAQKQGWKTLFSGLSIN 186
G F Y L+ R ++ A + A + G + G L+ G +I+
Sbjct: 631 GASSLFFVYSLDYARTRLANDAKAAKKGAGGRQFNGLIDVYKKTLATDGIAGLYRGFNIS 810
Query: 187 YIKVVPSAAIGFTVYDTMKSYL 208
+ ++ + F +YD++K L
Sbjct: 811 CVGIIVYRGLYFGMYDSLKPVL 876
>TC87378 similar to PIR|S21313|S21313 ADP ATP carrier protein - Arabidopsis
thaliana (fragment), partial (54%)
Length = 1109
Score = 70.5 bits (171), Expect = 5e-13
Identities = 44/184 (23%), Positives = 84/184 (44%), Gaps = 7/184 (3%)
Frame = +1
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTY 74
FA + L GG++ +K+ AP+ER+K+L Q + ++ +G ++ + Y+GI DC +T
Sbjct: 523 FATDFLMGGVSAAVSKSAAAPIERVKLLIQ--NQDEMIKAGRLS--EPYKGIGDCFGRTV 690
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMK-----RHVPEDYKKSIMAKLTCGSVA 129
K+ G ++RG + FP F F + K + + Y K + G+ A
Sbjct: 691 KDEGFASLWRGNTVNVIRYFPTQAFNFAFKDYFKKLFNFKQERDGYWKVFAGNIASGAAA 870
Query: 130 GLLGQTFTYFLEVVRRQM--QVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINY 187
G F Y L+ R ++ ++ E + G + Q G L+ G +++
Sbjct: 871 GASSSIFVYSLDYARTRLASDAKSTKKGGERQFNGLIDVYKKTLQSDGIAGLYRGFNVSV 1050
Query: 188 IKVV 191
+ ++
Sbjct: 1051VGII 1062
>BG644374 similar to GP|21536790|gb ADP/ATP translocase-like protein
{Arabidopsis thaliana}, partial (44%)
Length = 753
Score = 68.9 bits (167), Expect = 1e-12
Identities = 43/174 (24%), Positives = 83/174 (46%), Gaps = 5/174 (2%)
Frame = +2
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTY 74
F ++L+AG + GG T+VAP+ER K+L Q + N++ + + ++G+ DC+ T
Sbjct: 242 FNRDLMAGAVMGGAVHTIVAPIERAKLLLQT---QESNIAILSGPHRRFKGMLDCIGSTV 412
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYK-----KSIMAKLTCGSVA 129
KE G+ ++RG ++ +P L F + + + +++ A GS A
Sbjct: 413 KEEGVLSLWRGNGSSVLRYYPSVALNFSLKDLYRNILHSNFQDGHFLAGSSANFIAGSAA 592
Query: 130 GLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGL 183
G Y L++ ++ +L E + +G + I +K G + ++ GL
Sbjct: 593 GCTTLIIIYPLDISHTRL-AADLGRYETRQFRGIYHFLRTIHEKDGIRGIYRGL 751
>TC81666 similar to PIR|T07793|T07793 uncoupling protein (clone StUCP7)
mitochonrial - potato, partial (85%)
Length = 911
Score = 65.1 bits (157), Expect = 2e-11
Identities = 57/220 (25%), Positives = 103/220 (45%), Gaps = 9/220 (4%)
Frame = +3
Query: 1 MAASSESILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNE 60
M A S+S L P FA + + FA+ PL+ K+ Q+ K V+G V++
Sbjct: 114 MVADSKSNLSFGPTFA----SSAFSACFAEVCTIPLDTAKVRLQL---QKQAVAGDVSSL 272
Query: 61 QVYRGIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR------HVPE- 113
Y+G+ + +E G+ +++G+ P L Y GL+ YE +K HV +
Sbjct: 273 PKYKGMLGTVGTIAREEGLSALWKGIVPGLHRQCLYGGLRIGLYEPVKTFYTGSDHVGDV 452
Query: 114 DYKKSIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQ-NLPASEEAELKGTMRSMVLIAQ 172
K I+A T G+VA ++ ++V+ ++Q + LP G++ + I +
Sbjct: 453 PLSKKILAAFTTGAVAIMVANP----TDLVKVRLQAEGKLPPGVPRRYSGSLNAYSSIVR 620
Query: 173 KQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMK-SYLRVP 211
++G + L++GL N + A YD +K + L++P
Sbjct: 621 QEGVRALWTGLGPNIARNGIINAAELASYDQVKQTILKIP 740
>TC92386 similar to GP|10177187|dbj|BAB10321. mitochondrial carrier
protein-like {Arabidopsis thaliana}, partial (56%)
Length = 813
Score = 63.9 bits (154), Expect = 4e-11
Identities = 47/174 (27%), Positives = 82/174 (47%), Gaps = 8/174 (4%)
Frame = +2
Query: 21 AGGLAGGFAKTVVAPLERLKILFQV-VSPTK----LNVSGMVNNEQVYRGIRDCLSKTYK 75
AG ++GG ++TV +PL+ +KI FQV + PT L + + Y G+ +
Sbjct: 233 AGAISGGISRTVTSPLDVIKIRFQVQLEPTSSWALLQKDLVSSAPSKYTGMLQATKDILR 412
Query: 76 EGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLT--CGSVAGLLG 133
E G+KG +RG P L + PY ++F ++K K L+ V+G L
Sbjct: 413 EEGLKGFWRGNVPALLMVMPYTAIQFTVLHKLKTFASGSSKSENHTNLSPYLSYVSGALS 592
Query: 134 QTFTYFLEVVRRQMQVQNLPASEEAELKGTMRS-MVLIAQKQGWKTLFSGLSIN 186
+ +Y L + + L + + ++ MRS V I Q G++ +++G+ N
Sbjct: 593 RVCSYLLVHILFDLLRTILASQGDPKVYPNMRSAFVDIIQTPGFQGMYAGIVSN 754
Score = 31.2 bits (69), Expect = 0.32
Identities = 14/29 (48%), Positives = 18/29 (61%)
Frame = +3
Query: 80 KGIYRGVAPTLFGIFPYAGLKFYFYEEMK 108
K G++PTL I PYAGL+F Y+ K
Sbjct: 726 KACMLGLSPTLVEIIPYAGLQFGTYDTFK 812
Score = 29.3 bits (64), Expect = 1.2
Identities = 16/53 (30%), Positives = 27/53 (50%), Gaps = 7/53 (13%)
Frame = +3
Query: 160 LKGTMRSMV-------LIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMK 205
LK T R ++ + ++ Q +K GLS ++++P A + F YDT K
Sbjct: 654 LKATQRYILT*GQHSSISSRLQDFKACMLGLSPTLVEIIPYAGLQFGTYDTFK 812
>BG644332 SP|P27081|ADT2 ADP ATP carrier protein mitochondrial precursor
(ADP/ATP translocase), partial (56%)
Length = 749
Score = 62.8 bits (151), Expect = 1e-10
Identities = 41/139 (29%), Positives = 67/139 (47%), Gaps = 6/139 (4%)
Frame = +2
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTY 74
FA + L GG++ +KT AP+ER+K+L Q + ++ +G ++ + Y+GI DC S+T
Sbjct: 338 FATDFLMGGVSAAVSKTAAAPIERVKLLIQ--NQDEMIKAGRLS--EPYKGIGDCFSRTI 505
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLTC------GSV 128
K+ G ++RG + FP L F F + KR + K+ C V
Sbjct: 506 KDEGFAALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDRDGYWKVVCR*PGIWWVV 685
Query: 129 AGLLGQTFTYFLEVVRRQM 147
G F Y L+ R ++
Sbjct: 686 PGASSLLFVYSLDYARTRL 742
>BF635367 similar to PIR|T01729|T01 mitochondrial solute carrier protein
homolog - Arabidopsis thaliana, partial (30%)
Length = 584
Score = 62.4 bits (150), Expect = 1e-10
Identities = 35/100 (35%), Positives = 53/100 (53%)
Frame = +1
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 76
K L+AGG+AGG ++T VAPLERLKIL QV + + +G V L +K
Sbjct: 217 KSLVAGGVAGGVSRTAVAPLERLKILLQVQNRHNVKYNGTVQG----------LKYIWKT 366
Query: 77 GGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYK 116
G +G+++G I P + +KF+ YE+ + Y+
Sbjct: 367 EGFRGMFKGNGTNCARIIPNSAVKFFSYEQASNGILSLYR 486
Score = 43.5 bits (101), Expect = 6e-05
Identities = 24/85 (28%), Positives = 46/85 (53%)
Frame = +1
Query: 118 SIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWK 177
+I L G VAG + +T LE ++ +QVQN + GT++ + I + +G++
Sbjct: 208 TICKSLVAGGVAGGVSRTAVAPLERLKILLQVQN---RHNVKYNGTVQGLKYIWKTEGFR 378
Query: 178 TLFSGLSINYIKVVPSAAIGFTVYD 202
+F G N +++P++A+ F Y+
Sbjct: 379 GMFKGNGTNCARIIPNSAVKFFSYE 453
>TC91099 similar to GP|10177844|dbj|BAB11273. ADP/ATP translocase-like
protein {Arabidopsis thaliana}, partial (39%)
Length = 802
Score = 59.3 bits (142), Expect = 1e-09
Identities = 27/87 (31%), Positives = 52/87 (59%)
Frame = +3
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTY 74
F ++L+AG + GG T+VAP+ER K+L Q + N++ + + + ++G+ DC+ +T
Sbjct: 378 FQRDLMAGAVMGGAVHTIVAPIERAKLLLQT---QESNLAIVASGRRKFKGMFDCIIRTV 548
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKF 101
+E G+ ++RG ++ +P L F
Sbjct: 549 REEGVISLWRGNGSSVLRYYPSVALNF 629
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.138 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,195,123
Number of Sequences: 36976
Number of extensions: 58006
Number of successful extensions: 316
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 266
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 287
length of query: 217
length of database: 9,014,727
effective HSP length: 92
effective length of query: 125
effective length of database: 5,612,935
effective search space: 701616875
effective search space used: 701616875
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC148360.13


