

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148359.9 + phase: 0
(877 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
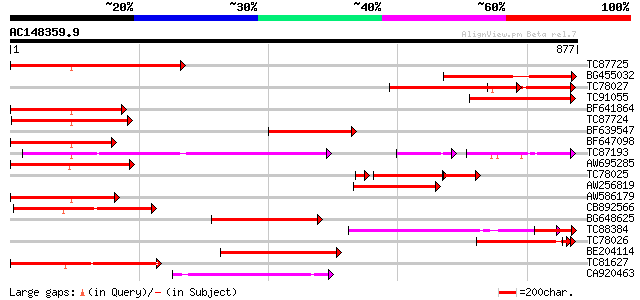
Score E
Sequences producing significant alignments: (bits) Value
TC87725 similar to GP|5817341|gb|AAD52714.1| putative NBS-LRR ty... 395 e-110
BG455032 weakly similar to GP|10440622|gb| putative NBS-LRR type... 280 2e-75
TC78027 similar to PIR|D86327|D86327 protein F18O14.17 [imported... 277 1e-74
TC91055 weakly similar to GP|10440622|gb|AAG16860.1 putative NBS... 261 6e-70
BF641864 243 3e-64
TC87724 241 9e-64
BF639547 similar to GP|15553678|gb I2C-5 {Lycopersicon pimpinell... 233 2e-61
BF647098 weakly similar to GP|22324956|gb| putative disease resi... 231 7e-61
TC87193 similar to GP|21616922|gb|AAM66423.1 NBS-LRR protein {Or... 231 9e-61
AW695285 similar to GP|15487944|gb| NBS/LRR resistance protein-l... 231 9e-61
TC78025 151 1e-58
AW256819 homologue to GP|7302697|gb|A CG5784-PA {Drosophila mela... 220 2e-57
AW586179 209 3e-54
CB892566 weakly similar to GP|15289776|db Lycopersicon esculentu... 204 9e-53
BG648625 weakly similar to GP|20385440|gb| resistance gene analo... 193 2e-49
TC88384 homologue to GP|21626661|gb|AAF47055.2 CG13560-PA {Droso... 193 3e-49
TC78026 homologue to PIR|T31434|T31434 densin-180 - rat, partial... 191 1e-48
BE204114 weakly similar to GP|15553678|gb I2C-5 {Lycopersicon pi... 180 2e-45
TC81627 weakly similar to GP|18652501|gb|AAL77135.1 Putative NBS... 176 3e-44
CA920463 weakly similar to GP|2852684|gb|A resistance protein ca... 175 8e-44
>TC87725 similar to GP|5817341|gb|AAD52714.1| putative NBS-LRR type disease
resistance protein {Pisum sativum}, partial (44%)
Length = 899
Score = 395 bits (1016), Expect = e-110
Identities = 213/277 (76%), Positives = 227/277 (81%), Gaps = 5/277 (1%)
Frame = +3
Query: 1 MADPFLGVVFENLMSLLQIEFSTIYGIKSKAENLSTTLVDIRAVLEDAEKRQVTDNFIKV 60
MAD LGVVFENL +L Q EFSTI GIKSKA+ LS LV I+AVLEDAEK+Q + IK+
Sbjct: 69 MADALLGVVFENLTALHQNEFSTISGIKSKAQKLSDNLVHIKAVLEDAEKKQFKELSIKL 248
Query: 61 WLQDLKDVVYVLDDILDECSIKSSRLKKFTSLK-----FRHKIGNRLKEITGRLDRIAER 115
WLQDLKD VYVLDDILDE SIKS +L+ TSLK FR++IGNRLKEIT RLD IAE
Sbjct: 249 WLQDLKDAVYVLDDILDEYSIKSGQLRGSTSLKPKNIKFRNEIGNRLKEITRRLDNIAES 428
Query: 116 KNKFSLQTGGTLRESPYQVAEGRQTSSTPLETKALGRDDDKEKIVEFLLTHAKDSDFISV 175
KNKFSLQ GGTLRE P QVAEGRQT S E K GR+ DKEKIVEFLLT AKDSDFISV
Sbjct: 429 KNKFSLQMGGTLREIPDQVAEGRQTGSIIAEPKVFGREVDKEKIVEFLLTQAKDSDFISV 608
Query: 176 YPIVGLGGIGKTTLVQLIYNDVRVSDNFDKKIWVCVSETFSVKRILCSIIESITLEKCPD 235
YPI GLGGIGKTTLVQLI+NDVRVS +FDKK+WVCVSETFSVKRILCSI ESITL KCPD
Sbjct: 609 YPIFGLGGIGKTTLVQLIFNDVRVSGHFDKKVWVCVSETFSVKRILCSIFESITLXKCPD 788
Query: 236 FELDVMERKVQGLLQGKIYLLILDDVWNQNEQLEYGL 272
FE VME KVQGL QGK YLL LDDVWNQNEQLE GL
Sbjct: 789 FEYAVMEGKVQGLXQGKRYLLXLDDVWNQNEQLESGL 899
>BG455032 weakly similar to GP|10440622|gb| putative NBS-LRR type resistance
protein 3' partial {Oryza sativa}, partial (3%)
Length = 666
Score = 280 bits (716), Expect = 2e-75
Identities = 142/206 (68%), Positives = 158/206 (75%)
Frame = +1
Query: 671 EVRVFPSLKVLHLYELPNIEGLLKVERGKVFPCLSRLTIYYCPKLGLPCLPSLKSLNVSG 730
EVR FPSL+VL L+ LPNIEGLLKVERG++FPCLS L I+ CPKLGLPCLPSLK L V G
Sbjct: 1 EVRAFPSLEVLELHGLPNIEGLLKVERGEMFPCLSSLDIWKCPKLGLPCLPSLKDLGVDG 180
Query: 731 CNNELLRSIPTFRGLTELTLYNGEGITSFPEGMFKNLTSLQSLFVDNFPNLKELPNEPFN 790
NNELLRSI TFRGLT+LTL +GEGITS PE MFKNLTSLQSLFV P
Sbjct: 181 RNNELLRSISTFRGLTQLTLNSGEGITSLPEEMFKNLTSLQSLFVTFLP----------- 327
Query: 791 PALTHLYIYNCNEIESLPEKMWEGLQSLRTLEIWDCKGMRCLPEGIRHLTSLEFLRIWSC 850
++ESLPE+ WEGLQSLR L IW C+G+RCLPEGIRHLTSLE L I C
Sbjct: 328 ------------QLESLPEQNWEGLQSLRALLIWGCRGLRCLPEGIRHLTSLELLSIIDC 471
Query: 851 PTLEERCKEGTGEDWDKIAHIPKIKI 876
PTL+ERCKEGTGEDWDKIAHIP+I++
Sbjct: 472 PTLKERCKEGTGEDWDKIAHIPRIEL 549
>TC78027 similar to PIR|D86327|D86327 protein F18O14.17 [imported] -
Arabidopsis thaliana, partial (6%)
Length = 2226
Score = 277 bits (708), Expect = 1e-74
Identities = 143/207 (69%), Positives = 167/207 (80%), Gaps = 3/207 (1%)
Frame = +1
Query: 588 VEQLLKVLQPHSNLKCLEIKYYDGLSLPSWVSILSNLVSLELGDCKKFVRLPLLGKLPSL 647
+ + L+VLQPHSNLKCL I YY+GLSLPSW+ ILSNL+SLEL C K VRLPLLGKLPSL
Sbjct: 28 LSKYLEVLQPHSNLKCLTINYYEGLSLPSWIIILSNLISLELEICNKIVRLPLLGKLPSL 207
Query: 648 EKLELSSMVNLKYLDDDESQDGMEVRVFPSLKVLHLYELPNIEGLLKVERGKVFPCLSRL 707
+KL L M NLKYLDDDES+ GMEV VFPSL+ L+L LPNIEGLLKVERG++FPCLS+L
Sbjct: 208 KKLRLYGMNNLKYLDDDESEYGMEVSVFPSLEELNLKSLPNIEGLLKVERGEMFPCLSKL 387
Query: 708 TIYYCPKLGLPCLPSLKSLNVSGCNNELLRSIPTFRGLTELTLYNGEGITSFPEGMFKNL 767
I+ CP+LGLPCLPSLKSL++ CNNELLRSI TFRGLT+LTL +GEGITS PE MFKNL
Sbjct: 388 DIWDCPELGLPCLPSLKSLHLWECNNELLRSISTFRGLTQLTLNSGEGITSLPEEMFKNL 567
Query: 768 TSLQ---SLFVDNFPNLKELPNEPFNP 791
TSL S+ V ++ + + FNP
Sbjct: 568 TSLHLCVSIVVMSWSPYQSKTGKVFNP 648
Score = 138 bits (348), Expect = 8e-33
Identities = 75/140 (53%), Positives = 91/140 (64%), Gaps = 4/140 (2%)
Frame = +3
Query: 739 IPTFRGL----TELTLYNGEGITSFPEGMFKNLTSLQSLFVDNFPNLKELPNEPFNPALT 794
IP F G+ TE+ LY +++P+ +N N ++ F+ +L
Sbjct: 438 IPPFMGM*Q*VTEVNLYLPWSYSAYPK*W*RN----------NILTRGDVQKPYFSSSLC 587
Query: 795 HLYIYNCNEIESLPEKMWEGLQSLRTLEIWDCKGMRCLPEGIRHLTSLEFLRIWSCPTLE 854
I CNE+ESLPE+ WEGLQSLR L+IW C+G+RCLPEGIRHLTSLE L I CPTLE
Sbjct: 588 ---INCCNELESLPEQNWEGLQSLRALQIWGCRGLRCLPEGIRHLTSLELLDIIDCPTLE 758
Query: 855 ERCKEGTGEDWDKIAHIPKI 874
ERCKEGT EDWDKIAHIPKI
Sbjct: 759 ERCKEGTWEDWDKIAHIPKI 818
>TC91055 weakly similar to GP|10440622|gb|AAG16860.1 putative NBS-LRR type
resistance protein 3' partial {Oryza sativa}, partial
(2%)
Length = 978
Score = 261 bits (668), Expect = 6e-70
Identities = 128/164 (78%), Positives = 140/164 (85%)
Frame = +3
Query: 712 CPKLGLPCLPSLKSLNVSGCNNELLRSIPTFRGLTELTLYNGEGITSFPEGMFKNLTSLQ 771
C KLGLPCLPSLKSL VS CNNELLRSI TFRGLT+L + GEGITSFPEGMFKNLTSLQ
Sbjct: 12 CRKLGLPCLPSLKSLTVSECNNELLRSISTFRGLTQLFVNGGEGITSFPEGMFKNLTSLQ 191
Query: 772 SLFVDNFPNLKELPNEPFNPALTHLYIYNCNEIESLPEKMWEGLQSLRTLEIWDCKGMRC 831
SL + NFP LKELPNE FNPALT L I CNE+ESLPE+ WEGLQSLRTL I+ C+G+RC
Sbjct: 192 SLRIYNFPKLKELPNETFNPALTLLCICYCNELESLPEQNWEGLQSLRTLHIYSCEGLRC 371
Query: 832 LPEGIRHLTSLEFLRIWSCPTLEERCKEGTGEDWDKIAHIPKIK 875
LPEGIRHLTSLE L I C TL+ERCK+ TGEDWDKI+HIPKI+
Sbjct: 372 LPEGIRHLTSLELLTIIGCRTLKERCKKRTGEDWDKISHIPKIQ 503
>BF641864
Length = 656
Score = 243 bits (619), Expect = 3e-64
Identities = 133/185 (71%), Positives = 144/185 (76%), Gaps = 5/185 (2%)
Frame = +1
Query: 1 MADPFLGVVFENLMSLLQIEFSTIYGIKSKAENLSTTLVDIRAVLEDAEKRQVTDNFIKV 60
MAD LGVV ENL SLLQ EF+TI GI+SKA LS LV I+AVLEDAEK+Q + IK
Sbjct: 100 MADALLGVVSENLTSLLQNEFATISGIRSKARKLSDNLVHIKAVLEDAEKKQFKELSIKQ 279
Query: 61 WLQDLKDVVYVLDDILDECSIKSSRLKKFTSLK-----FRHKIGNRLKEITGRLDRIAER 115
WLQDLKD VYVL DILDE SI+S RL+ F S K FRH+IG+R KEIT RLD IAE
Sbjct: 280 WLQDLKDAVYVLGDILDEYSIESGRLRGFNSFKPMNIAFRHEIGSRFKEITRRLDDIAES 459
Query: 116 KNKFSLQTGGTLRESPYQVAEGRQTSSTPLETKALGRDDDKEKIVEFLLTHAKDSDFISV 175
KNKFSLQ GGTLRE P QVAEGRQT STPLE+KALGRD DK+K FLLTHAKDSDFISV
Sbjct: 460 KNKFSLQMGGTLREIPXQVAEGRQTXSTPLESKALGRDXDKKKXXXFLLTHAKDSDFISV 639
Query: 176 YPIVG 180
YPI G
Sbjct: 640 YPIXG 654
>TC87724
Length = 720
Score = 241 bits (615), Expect = 9e-64
Identities = 135/191 (70%), Positives = 145/191 (75%), Gaps = 5/191 (2%)
Frame = +2
Query: 4 PFLGVVFENLMSLLQIEFSTIYGIKSKAENLSTTLVDIRAVLEDAEKRQVTDNFIKVWLQ 63
P LGVVFENL SLLQ EFSTI GIKSKA+ LS LV I+AVLEDAEK+Q + IK+WLQ
Sbjct: 146 PLLGVVFENLTSLLQNEFSTISGIKSKAQKLSDNLVRIKAVLEDAEKKQFKELSIKLWLQ 325
Query: 64 DLKDVVYVLDDILDECSIKSSRLKKFTSLK-----FRHKIGNRLKEITGRLDRIAERKNK 118
DLKD VYVLDDILDE SIKS RL+ TS K FRH+IGNRLKEIT RLD IAE KNK
Sbjct: 326 DLKDAVYVLDDILDEYSIKSCRLRGCTSFKPKNIMFRHEIGNRLKEITRRLDDIAESKNK 505
Query: 119 FSLQTGGTLRESPYQVAEGRQTSSTPLETKALGRDDDKEKIVEFLLTHAKDSDFISVYPI 178
FSLQ GGTLRE P QVAEGRQT S E K GR+ DKEKI EFLLT A+DSDF+SVYPI
Sbjct: 506 FSLQMGGTLREIPDQVAEGRQTGSIIAEPKVFGREVDKEKIAEFLLTQARDSDFLSVYPI 685
Query: 179 VGLGGIGKTTL 189
V K TL
Sbjct: 686 VWFXWCWKNTL 718
>BF639547 similar to GP|15553678|gb I2C-5 {Lycopersicon pimpinellifolium},
partial (4%)
Length = 660
Score = 233 bits (595), Expect = 2e-61
Identities = 115/137 (83%), Positives = 121/137 (87%), Gaps = 1/137 (0%)
Frame = +1
Query: 401 FYLTPTLKQCFSFCAIFPKDREILKEELIQLWMANGFIA-KRNLEVEDVGNMVWKELYQK 459
FYLTPTLKQCFSFCAIFPKD EILKEELIQLWMANGFI+ K NL+VEDVGNMVWKELYQK
Sbjct: 67 FYLTPTLKQCFSFCAIFPKDGEILKEELIQLWMANGFISSKGNLDVEDVGNMVWKELYQK 246
Query: 460 SFFQDCKMGEYSGDISFKMHDLIHDLAQSVMGQECMYLENANMSSLTKSTHHISFNSDTF 519
SFFQD KM EYSGDI FKMHDL+HDLAQSVMGQEC+YLENANM+SLTKSTHHISFNSD
Sbjct: 247 SFFQDIKMDEYSGDIFFKMHDLVHDLAQSVMGQECVYLENANMTSLTKSTHHISFNSDNL 426
Query: 520 LSFDEEKGNSLTELRDL 536
LSFDE + LR L
Sbjct: 427 LSFDEGAFKKVESLRTL 477
Score = 39.3 bits (90), Expect = 0.007
Identities = 20/32 (62%), Positives = 20/32 (62%)
Frame = +3
Query: 380 DSELWDLPQEKSILPALRLSYFYLTPTLKQCF 411
DSELW LPQE SIL ALRLSY K F
Sbjct: 3 DSELWALPQENSILLALRLSYLLFNTNPKTMF 98
>BF647098 weakly similar to GP|22324956|gb| putative disease resistant
protein {Oryza sativa (japonica cultivar-group)},
partial (2%)
Length = 683
Score = 231 bits (590), Expect = 7e-61
Identities = 129/170 (75%), Positives = 138/170 (80%), Gaps = 5/170 (2%)
Frame = +2
Query: 1 MADPFLGVVFENLMSLLQIEFSTIYGIKSKAENLSTTLVDIRAVLEDAEKRQVTDNFIKV 60
MA LGVVFENL SLLQ EFSTI GIKSKA+ LS LV I+AVLEDAEK+Q + IK+
Sbjct: 173 MACALLGVVFENLTSLLQNEFSTISGIKSKAQKLSDNLVHIKAVLEDAEKKQFKELSIKL 352
Query: 61 WLQDLKDVVYVLDDILDECSIKSSRLKKFTSLK-----FRHKIGNRLKEITGRLDRIAER 115
WLQDLKD VYVLDDILDE SI+S RL+ FTS K FRH+IGNRLKEIT RLD IAER
Sbjct: 353 WLQDLKDAVYVLDDILDEYSIESCRLRGFTSFKPKNIMFRHEIGNRLKEITRRLDDIAER 532
Query: 116 KNKFSLQTGGTLRESPYQVAEGRQTSSTPLETKALGRDDDKEKIVEFLLT 165
KNKFSLQTG TLR P QVAEGRQTSSTP E+KALGRDDD EKIVEFLLT
Sbjct: 533 KNKFSLQTGETLRVIPDQVAEGRQTSSTPXESKALGRDDDXEKIVEFLLT 682
>TC87193 similar to GP|21616922|gb|AAM66423.1 NBS-LRR protein {Oryza sativa
(japonica cultivar-group)}, partial (26%)
Length = 3119
Score = 231 bits (589), Expect = 9e-61
Identities = 153/501 (30%), Positives = 255/501 (50%), Gaps = 23/501 (4%)
Frame = +3
Query: 20 EFSTIYGIKSKAENLSTTLVDIRAVLEDAEKRQVTDNFIKVWLQDLKDVVYVLDDILDEC 79
E IYG+ + E L T+ I+AVL DAE +Q + +++W++ L D++ DD++DE
Sbjct: 147 EIGRIYGVMDELERLKKTVESIKAVLLDAEDKQEQSHAVQLWVRRLNDLLLPADDLIDEF 326
Query: 80 SIKSSRLKKFTSLK----------------FRHKIGNRLKEITGRLDRIAERKNKFSLQT 123
I+ K+ + K FR K+ + +++I + + + L +
Sbjct: 327 FIEDMIHKRDKAHKNKVKQVFHSFSPSRTAFRRKMAHEIEKIQRSFKDVEKDMSYLKLNS 506
Query: 124 GGTLRESPYQVAEGRQTSSTPLETKALGRDDDKEKIVEFLLTHAKDSDFISVYPIVGLGG 183
+ V R+T S LE++ +GR+DDK KI+ LL + + +S+ IVG+GG
Sbjct: 507 NVVVVAKTNNVR--RETCSYVLESEIIGREDDKNKIIS-LLRQSHEHQNVSLVAIVGIGG 677
Query: 184 IGKTTLVQLIYNDVRVSDNFDKKIWVCVSETFSVKRILCSIIESITLEKCPDFELDVMER 243
+GKT L QL+Y D V + F+K +WVCVS+ F K IL +++ S+T E + L ++
Sbjct: 678 LGKTALAQLVYKDGEVKNLFEKHMWVCVSDNFDFKTILKNMVASLTKEDVVNKTLQELQS 857
Query: 244 KVQGLLQGKIYLLILDDVWNQNEQLEYGLTQDRWNRLKSVLSCGSKGSSILVSTRDKDVA 303
+Q L GK YLL+LDDVWN+ + ++W++L+ L CG++GS ++++T K VA
Sbjct: 858 MLQVNLTGKRYLLVLDDVWNE--------SFEKWDQLRPYLMCGAQGSKVVMTTCSKIVA 1013
Query: 304 TIMGTCQAHSLSGLSDSDCWLLFKQHAFRHYR-EEHTKLVEIGKEIVKKCNGLPLAAKAL 362
MG H L GL+ W+LFK F + L IGK+I +KC G+PLA ++L
Sbjct: 1014DRMGVSDQHVLRGLTPEKSWVLFKNIVFGDVTVGVNQPLESIGKKIAEKCKGVPLAIRSL 1193
Query: 363 GGLMFSMNEEKEWLDIKDSELWD--LPQEKSILPALRLSYFYLTPTLKQCFSFCAIFPKD 420
GG++ S ++E EW+++ E + + +E SI+ ++ L + K F
Sbjct: 1194GGILRSESKESEWINVLQGECLENCVIRENSIIAGPKIELPELITSTKAMFCLLLFISSG 1373
Query: 421 REILK---EELIQLWMANGFIAKRNLEVEDVGNMVWKELYQKSFFQDCKMG-EYSGDISF 476
I + + + + + + E + K ++ CK + F
Sbjct: 1374LGI*EG*VNSNVDGTRLSWLFS*KAMH-ERCW*SICKNFLEEFILPRCKFE*SWVMVTGF 1550
Query: 477 KMHDLIHDLAQSVMGQECMYL 497
KMHDL+HDLA V G + + L
Sbjct: 1551KMHDLMHDLATQVAGNDLLVL 1613
Score = 64.3 bits (155), Expect(2) = 3e-20
Identities = 57/192 (29%), Positives = 92/192 (47%), Gaps = 24/192 (12%)
Frame = +1
Query: 707 LTIYYCPKLG-LPCLPSLKS-LNVSGCNNELLRSIPTFR------GLTELTLYNG----- 753
L I C L +P P++K L + C+ E+L + G L++
Sbjct: 2398 LVIVRCKMLTFMPTFPNIKKKLELWSCSAEILEATLNIAESQYSIGFPPLSILKSFHIIE 2577
Query: 754 --EGITSFPEGMFKNLTSLQSLFVDNFPNLK-ELPNEPFN------PALTHLYIYNCNEI 804
G+ + P+ KNLTSL++L +D+ + + E+ F P+L + I C +
Sbjct: 2578 TIMGMENVPKDWLKNLTSLENLDLDSLSSQQFEVIEMWFKDDLICLPSLQKIQIRTCR-L 2754
Query: 805 ESLPEKMWEGLQSLRTLEIWDCKGMRCLPEGIRHLTSLEFLRIWSCPT--LEERCKEGTG 862
++LP+ + + SL+ L ++ C+ + LPEG+ LT+L L I CP + TG
Sbjct: 2755 KALPDWICN-ISSLQHLAVYGCESLVDLPEGMPRLTNLHTLEIIGCPISYYYDEFLRKTG 2931
Query: 863 EDWDKIAHIPKI 874
W KIAHIPKI
Sbjct: 2932 ATWSKIAHIPKI 2967
Score = 53.1 bits (126), Expect(2) = 3e-20
Identities = 31/93 (33%), Positives = 55/93 (58%)
Frame = +2
Query: 599 SNLKCLEIKYYDGLSLPSWVSILSNLVSLELGDCKKFVRLPLLGKLPSLEKLELSSMVNL 658
S + L I+ GLSL +W+S ++N++ L L +C+ LP L +LP L+ L+L + L
Sbjct: 2045 SGFRKLSIQQPKGLSLSNWLSPVTNIIELCLYNCQDLRYLPPLERLPFLKSLDLRWLNQL 2224
Query: 659 KYLDDDESQDGMEVRVFPSLKVLHLYELPNIEG 691
+Y+ +E + FPSL++L+ +E ++G
Sbjct: 2225 EYIYYEEPI--LHESFFPSLEILNFFECWKLKG 2317
>AW695285 similar to GP|15487944|gb| NBS/LRR resistance protein-like protein
{Theobroma cacao}, partial (9%)
Length = 618
Score = 231 bits (589), Expect = 9e-61
Identities = 131/198 (66%), Positives = 150/198 (75%), Gaps = 6/198 (3%)
Frame = +2
Query: 1 MADPFLGVVFENLMSLLQIEFSTIYGIKSKAENLSTTLVDIRAVLEDAEKRQVTDNFIKV 60
MA+ L FE + SLLQ EFSTI GIKSKA+NLST+L I AVL DAEKRQV D++IKV
Sbjct: 23 MAEALLRAAFEKVNSLLQSEFSTISGIKSKAKNLSTSLNHIEAVLVDAEKRQVKDSYIKV 202
Query: 61 WLQDLKDVVYVLDDILDECSIKSSRLKKFTS-----LKFRHKIGNRLKEITGRLDRIAER 115
WLQ LKD VYVLDDILDECSI+S+RL S + FR +IGNRLKEIT RLD IA+
Sbjct: 203 WLQQLKDAVYVLDDILDECSIESARLGGSFSFNPKNIVFRRQIGNRLKEITRRLDDIADI 382
Query: 116 KNKFSLQTGGT-LRESPYQVAEGRQTSSTPLETKALGRDDDKEKIVEFLLTHAKDSDFIS 174
KNKF L+ G +RES +V E RQ +S + + GR DDKEKI EFLLTHA+DSDF+S
Sbjct: 383 KNKFLLRDGTVYVRESSDEVDEWRQINSIIAKPEVFGRKDDKEKIFEFLLTHARDSDFLS 562
Query: 175 VYPIVGLGGIGKTTLVQL 192
VYPIVGLGGIGKTTLVQL
Sbjct: 563 VYPIVGLGGIGKTTLVQL 616
>TC78025
Length = 1947
Score = 151 bits (381), Expect(3) = 1e-58
Identities = 75/113 (66%), Positives = 85/113 (74%)
Frame = +2
Query: 564 KKNLEKLCLSWENNDGFTKPPTISVEQLLKVLQPHSNLKCLEIKYYDGLSLPSWVSILSN 623
KK LSW++ G K P +S EQ+L+ LQPHSNLKCL I YY+GLSLPSW+ ILSN
Sbjct: 92 KKTFMNYTLSWKDKQGIPKTPVVSAEQVLEELQPHSNLKCLTINYYEGLSLPSWIIILSN 271
Query: 624 LVSLELGDCKKFVRLPLLGKLPSLEKLELSSMVNLKYLDDDESQDGMEVRVFP 676
LVSL L CKK VRLPLLGKLPSL+KL L + NLKYLDDDES+DGME FP
Sbjct: 272 LVSLVLLHCKKIVRLPLLGKLPSLKKLRLYGINNLKYLDDDESEDGMEGXGFP 430
Score = 78.2 bits (191), Expect(3) = 1e-58
Identities = 40/57 (70%), Positives = 45/57 (78%)
Frame = +3
Query: 672 VRVFPSLKVLHLYELPNIEGLLKVERGKVFPCLSRLTIYYCPKLGLPCLPSLKSLNV 728
V VFPSL++L L L NI GLLKVERG++FP LS+L I CPKLGLPCLPSLK L V
Sbjct: 417 VXVFPSLEILELSCLRNIVGLLKVERGEMFPSLSKLVIDCCPKLGLPCLPSLKDLYV 587
Score = 37.7 bits (86), Expect(3) = 1e-58
Identities = 18/22 (81%), Positives = 20/22 (90%)
Frame = +3
Query: 535 DLNLGGKLSIEGLKDVGSLSEA 556
DLNLGGKLSI+GL +VGSL EA
Sbjct: 3 DLNLGGKLSIKGLNNVGSLFEA 68
>AW256819 homologue to GP|7302697|gb|A CG5784-PA {Drosophila melanogaster},
partial (4%)
Length = 403
Score = 220 bits (560), Expect = 2e-57
Identities = 108/134 (80%), Positives = 121/134 (89%)
Frame = +1
Query: 533 LRDLNLGGKLSIEGLKDVGSLSEAQEANLMGKKNLEKLCLSWENNDGFTKPPTISVEQLL 592
LRDLNLGGKL IEGLKD GSLS+AQ A+LMGKK+L +LCLSWE+N GFT PPTIS +Q+L
Sbjct: 1 LRDLNLGGKLRIEGLKDFGSLSQAQAADLMGKKDLHELCLSWESNYGFTNPPTISAQQVL 180
Query: 593 KVLQPHSNLKCLEIKYYDGLSLPSWVSILSNLVSLELGDCKKFVRLPLLGKLPSLEKLEL 652
+VLQPHSNLKCL+I YYDGLSLPSW+ ILSNLVSLELG+CKK VRL L+GKLPSL+KLEL
Sbjct: 181 EVLQPHSNLKCLKINYYDGLSLPSWIIILSNLVSLELGNCKKVVRLQLIGKLPSLKKLEL 360
Query: 653 SSMVNLKYLDDDES 666
S M NLKYLDDDES
Sbjct: 361 SDMDNLKYLDDDES 402
>AW586179
Length = 590
Score = 209 bits (533), Expect = 3e-54
Identities = 115/175 (65%), Positives = 130/175 (73%), Gaps = 5/175 (2%)
Frame = +1
Query: 1 MADPFLGVVFENLMSLLQIEFSTIYGIKSKAENLSTTLVDIRAVLEDAEKRQVTDNFIKV 60
M D LG+V+ENL ++L E+STI GI K + LS LV I+AVLEDAEK+ + IK+
Sbjct: 64 MDDALLGLVYENLTAILHKEYSTISGINLKVQKLSNNLVHIKAVLEDAEKKHFKELSIKL 243
Query: 61 WLQDLKDVVYVLDDILDECSIKSSRLKKFTSLK-----FRHKIGNRLKEITGRLDRIAER 115
WLQDLKD VYVLDDILDE SIKS RL+ FTS K FRH+IGN KEIT RLD IAE
Sbjct: 244 WLQDLKDGVYVLDDILDEYSIKSCRLRGFTSFKPKNIMFRHEIGNTFKEITRRLDDIAES 423
Query: 116 KNKFSLQTGGTLRESPYQVAEGRQTSSTPLETKALGRDDDKEKIVEFLLTHAKDS 170
KNKFSLQ GGTLRE P+QVAEGRQT S E K GR+ DKEKIVEFLLT A+DS
Sbjct: 424 KNKFSLQMGGTLREIPHQVAEGRQTGSIIAEPKVFGREVDKEKIVEFLLTQARDS 588
>CB892566 weakly similar to GP|15289776|db Lycopersicon esculentum resistance
complex protein I2C-2 like protein, partial (1%)
Length = 688
Score = 204 bits (520), Expect = 9e-53
Identities = 116/231 (50%), Positives = 150/231 (64%), Gaps = 9/231 (3%)
Frame = +2
Query: 6 LGVVFENLMSLLQIEFSTIYGIKSKAENLSTTLVDIRAVLEDAEKRQVTDNFIKVWLQDL 65
LG++ +NL S +Q E +T G+ ++LS L IRAVL+DAEK+Q+T++ +K WLQ L
Sbjct: 5 LGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKEWLQQL 184
Query: 66 KDVVYVLDDILDECSI------KSSRLKKFTSLKF--RHKIGNRLKEITGRLDRIAERKN 117
+D YVLDDILDECSI + R+ +F +K R IG R+KEI +D IAE +
Sbjct: 185 RDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDIAEERM 364
Query: 118 KFSLQTGGTLRESPYQVAEGR-QTSSTPLETKALGRDDDKEKIVEFLLTHAKDSDFISVY 176
KF L G R+ EGR QT+S E+K GRD DKE IVEFLL HA DS+ +SVY
Sbjct: 365 KFGLHVGVIERQPE---DEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELSVY 535
Query: 177 PIVGLGGIGKTTLVQLIYNDVRVSDNFDKKIWVCVSETFSVKRILCSIIES 227
IVG GG GKTTL Q ++ND RV +FD KIWVCVS + ++L SIIE+
Sbjct: 536 SIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIEN 688
>BG648625 weakly similar to GP|20385440|gb| resistance gene analog {Vitis
vinifera}, partial (21%)
Length = 521
Score = 193 bits (491), Expect = 2e-49
Identities = 95/174 (54%), Positives = 125/174 (71%), Gaps = 1/174 (0%)
Frame = +3
Query: 312 HSLSGLSDSDCWLLFKQHAFRHYREEHTKLVEIGKEIVKKCNGLPLAAKALGGLMFSMNE 371
H L+ LSD D LFKQHAF RE +LVEIG+++V+KC G PLAAK LG L+ ++
Sbjct: 3 HPLAQLSDDDI*SLFKQHAFGANREGRAELVEIGQKLVRKCVGSPLAAKVLGSLLRFKSD 182
Query: 372 EKEWLDIKDSELWDLPQEKSILPALRLSYFYLTPTLKQCFSFCAIFPKDREILKEELIQL 431
E +W + +SE W+L + ++ ALRLSYF L +L+ CF+FCA+FPKD E+ KE IQL
Sbjct: 183 EHQWTSVVESEFWNLADDNHVMSALRLSYFNLKLSLRPCFTFCAVFPKDFEMEKEFFIQL 362
Query: 432 WMANGFIAKR-NLEVEDVGNMVWKELYQKSFFQDCKMGEYSGDISFKMHDLIHD 484
WMANG + R NL++E VGN VW ELYQ+SFFQ+ K + G+I+FKMHDL+HD
Sbjct: 363 WMANGLVTSRGNLQMEHVGNEVWNELYQRSFFQEIK-SDLVGNITFKMHDLVHD 521
>TC88384 homologue to GP|21626661|gb|AAF47055.2 CG13560-PA {Drosophila
melanogaster}, partial (3%)
Length = 1570
Score = 193 bits (490), Expect = 3e-49
Identities = 132/333 (39%), Positives = 184/333 (54%), Gaps = 4/333 (1%)
Frame = -3
Query: 525 EKGNSLTELRDLNLGGKLSIEGLKDVGSLSEAQEANLMGKKNLEKLCLSWENNDGFTKPP 584
++G L EL DL LGG+L I+GL++V S +A+EANL+GKK L +L LSW ++
Sbjct: 1385 KEGFGLAELNDLQLGGRLHIKGLENVSSEWDAKEANLIGKKELNRLYLSWGSHANSQGID 1206
Query: 585 TISVEQLLKVLQPHSNLKCLEIKYYDGLSLPSWV---SILSNLVSLELGDCKKFVRLPLL 641
T VEQ+L+ L+PH+ LK I+ Y G+ P W+ SIL LV + +C RLP L
Sbjct: 1205 T-DVEQVLEALEPHTGLKGFGIEGYVGIHFPHWMRNASILEGLVDITFYNCNNCQRLPPL 1029
Query: 642 GKLPSLEKLELSSMVNLKYLDDDESQDGMEVRVFPSLKVLHLYELPNIEGLLKVERGKVF 701
GKLP L L + M +LKY+DDD + R F SLK L L LPN+E +LK E ++
Sbjct: 1028 GKLPCLTTLYVFGMRDLKYIDDD-IYESTSKRAFISLKNLTLLGLPNLERMLKAEGVEML 852
Query: 702 PCLSRLTIYYCPKLGLPCLPSLKSLNVSGCNNELLRSIPTFRGLTELTLYNGEGITSFPE 761
P LS I PKL LP LPS++ L+V R +P ++ ++ FPE
Sbjct: 851 PQLSYFNISNVPKLALPSLPSIELLDV---GENKYRFLPQYK-----------VVSLFPE 714
Query: 762 GMFKNLTSLQSLFVDNFPNLKELPNE-PFNPALTHLYIYNCNEIESLPEKMWEGLQSLRT 820
+ ++ +L+ L + NF LK LP++ L L+I C+E+ES +G+ SLR
Sbjct: 713 RIVCSMHNLKLLIIGNFHKLKVLPDDLHCLSVLEELHISRCDELESFSMHALQGMISLRV 534
Query: 821 LEIWDCKGMRCLPEGIRHLTSLEFLRIWSCPTL 853
L I C + L EG+ L SLE L I C L
Sbjct: 533 LTIDLCGKLISLSEGMGDLASLERLVIHGCSQL 435
Score = 60.1 bits (144), Expect = 4e-09
Identities = 26/65 (40%), Positives = 39/65 (60%)
Frame = -2
Query: 812 WEGLQSLRTLEIWDCKGMRCLPEGIRHLTSLEFLRIWSCPTLEERCKEGTGEDWDKIAHI 871
WE + + C + LP ++L +L L I C LE+RCK+GTGEDW KIAH+
Sbjct: 282 WEP*LLFKE*KFISCTNAKSLPNSFQNLINLHTLLIVGCSKLEKRCKKGTGEDWQKIAHV 103
Query: 872 PKIKI 876
P++++
Sbjct: 102 PELEL 88
Score = 29.3 bits (64), Expect = 6.8
Identities = 15/32 (46%), Positives = 21/32 (64%)
Frame = -3
Query: 793 LTHLYIYNCNEIESLPEKMWEGLQSLRTLEIW 824
L HL I +CN + S+P K+ L SL+TL I+
Sbjct: 1490 LRHLVIKDCNSLYSMPSKI-SKLTSLKTLSIF 1398
>TC78026 homologue to PIR|T31434|T31434 densin-180 - rat, partial (1%)
Length = 828
Score = 191 bits (485), Expect = 1e-48
Identities = 96/146 (65%), Positives = 112/146 (75%)
Frame = +2
Query: 723 LKSLNVSGCNNELLRSIPTFRGLTELTLYNGEGITSFPEGMFKNLTSLQSLFVDNFPNLK 782
LK+ LRSI TFRGLT+L+LYNG GITSFPEGMFKNLTSLQSL V+ FP LK
Sbjct: 20 LKNSRYGDVTMSYLRSISTFRGLTQLSLYNGFGITSFPEGMFKNLTSLQSLSVNGFPKLK 199
Query: 783 ELPNEPFNPALTHLYIYNCNEIESLPEKMWEGLQSLRTLEIWDCKGMRCLPEGIRHLTSL 842
ELPNEPFNPALTHL I CNE+ESLPE+ WEGLQSLRTL+I +C+G+RCLPEGIRHLTSL
Sbjct: 200 ELPNEPFNPALTHLCITYCNELESLPEQNWEGLQSLRTLKIRNCEGLRCLPEGIRHLTSL 379
Query: 843 EFLRIWSCPTLEERCKEGTGEDWDKI 868
E+L + C++G +W +I
Sbjct: 380 EYL------*MSNICEKGAKREWGRI 439
Score = 44.3 bits (103), Expect = 2e-04
Identities = 18/21 (85%), Positives = 19/21 (89%)
Frame = +1
Query: 855 ERCKEGTGEDWDKIAHIPKIK 875
E CKEG GEDWDKIAHIPKI+
Sbjct: 409 EGCKEGMGEDWDKIAHIPKIQ 471
Score = 39.7 bits (91), Expect = 0.005
Identities = 22/50 (44%), Positives = 27/50 (54%)
Frame = +1
Query: 717 LPCLPSLKSLNVSGCNNELLRSIPTFRGLTELTLYNGEGITSFPEGMFKN 766
LPCLPSLK L + GCNNE L E+ LY ++FP F+N
Sbjct: 1 LPCLPSLKELEIWGCNNE----------LPEVNLYLPWSYSAFPI*RFRN 120
>BE204114 weakly similar to GP|15553678|gb I2C-5 {Lycopersicon
pimpinellifolium}, partial (3%)
Length = 583
Score = 180 bits (456), Expect = 2e-45
Identities = 93/187 (49%), Positives = 124/187 (65%), Gaps = 1/187 (0%)
Frame = +3
Query: 327 KQHAFRHYREEHTKLVEIGKEIVKKCNGLPLAAKALGGLMFSMNEEKEWLDIKDSELWDL 386
+Q AF E KLV IGKEI+KKC +PLAA ALG L+ EEKEWL +K+S+LW L
Sbjct: 3 RQRAFGPNEAEDEKLVVIGKEILKKCV*VPLAAIALGSLLRFKREEKEWLYVKESKLWSL 182
Query: 387 PQEKSILPALRLSYFYLTPTLKQCFSFCAIFPKDREILKEELIQLWMANGFIAKRN-LEV 445
E + AL+LSY L L+QCFSFCA+FPKD + K +I+LW+ANGFI+ L+
Sbjct: 183 EGEDYVKSALKLSYLNLPVKLRQCFSFCALFPKDEIMSKHFMIELWIANGFISSNQMLDA 362
Query: 446 EDVGNMVWKELYQKSFFQDCKMGEYSGDISFKMHDLIHDLAQSVMGQECMYLENANMSSL 505
E VGN VW ELY +SFFQD + E+ SFKMHDL+H+LA+SV + N ++ ++
Sbjct: 363 EGVGNEVWNELYWRSFFQDTETDEFGQITSFKMHDLVHELAESVTREVWCITYNNDLPTV 542
Query: 506 TKSTHHI 512
++S H+
Sbjct: 543 SESIRHL 563
>TC81627 weakly similar to GP|18652501|gb|AAL77135.1 Putative NBS-LRR type
resistance protein {Oryza sativa}, partial (6%)
Length = 890
Score = 176 bits (447), Expect = 3e-44
Identities = 106/242 (43%), Positives = 149/242 (60%), Gaps = 8/242 (3%)
Frame = +2
Query: 1 MADPFLGVVFENLMSLLQIEFSTIYGIKSKAENLSTTLVDIRAVLEDAEKRQVTDNFIKV 60
M D LG+V ENL+S ++ E ST G+ + L L IRAVL+DAE++Q+T +K
Sbjct: 146 MTDVLLGIVIENLISFVREELSTFLGVGELTQKLCGNLTAIRAVLQDAEEKQITSRAVKD 325
Query: 61 WLQDLKDVVYVLDDILDECSIKSSR------LKKFTSLKF--RHKIGNRLKEITGRLDRI 112
WLQ L D +VLDDILD+CS S + +F K + IG R+KE+ ++D I
Sbjct: 326 WLQKLADAAHVLDDILDDCSTTSKAHGDNKWIARFHPKKILAQRAIGKRMKEVAKKIDEI 505
Query: 113 AERKNKFSLQTGGTLRESPYQVAEGRQTSSTPLETKALGRDDDKEKIVEFLLTHAKDSDF 172
AE + K+ LQ T E E RQT+S E K GRD D EKIVE L++HA DS+
Sbjct: 506 AEGRIKYGLQVKVT-EEHQRGDDEWRQTTSVITEPKVYGRDHDIEKIVEVLVSHAIDSEE 682
Query: 173 ISVYPIVGLGGIGKTTLVQLIYNDVRVSDNFDKKIWVCVSETFSVKRILCSIIESITLEK 232
+SVY IVG+GG+GKTTL Q+++ND R+ +++ + +S + S+ +IL SII+S T+ K
Sbjct: 683 LSVYSIVGVGGLGKTTLAQVVFNDERLDTLYERLTNMKLSSSSSMMKILESIIKS-TVGK 859
Query: 233 CP 234
P
Sbjct: 860 NP 865
>CA920463 weakly similar to GP|2852684|gb|A resistance protein candidate
{Lactuca sativa}, partial (6%)
Length = 810
Score = 175 bits (443), Expect = 8e-44
Identities = 104/255 (40%), Positives = 153/255 (59%), Gaps = 5/255 (1%)
Frame = -1
Query: 252 KIYLLILDDVWNQNEQLEYGLTQDRWNRLKSVLSCGSKGSSILVSTRDKDVATIMGTCQA 311
K +LL LDDVW+++ +W +K++L G +GS +LV+TR +A +M T +
Sbjct: 756 KKFLLRLDDVWSEDRV--------KWIEVKNLLQVGDEGSKVLVTTRSHSIAKMMCTNTS 601
Query: 312 HSLSGLSDSDCWLLFKQHAFRHYREE-HTKLVEIGKEIVKKCNGLPLAAKALGGLMFSMN 370
++L GLS D +F + AF+ E+ + KL+EIGKEIV+KC GLPLA + LG +F +
Sbjct: 600 YTLQGLSREDSLSVFVKWAFKEGEEKKYPKLIEIGKEIVQKCGGLPLALRTLGSSLFLKD 421
Query: 371 EEKEWLDIKDSELWDLPQ-EKSILPALRLSYFYLTPTLKQCFSFCAIFPKDREILKEELI 429
+ +EW ++D+E+W+LPQ E ILPAL+LS+ L LK+CF+ ++F KD +
Sbjct: 420 DIEEWKFVRDNEIWNLPQKEDDILPALKLSFDQLPSYLKRCFACFSLFVKDFHFSNYSVT 241
Query: 430 QLWMANGFIAKRN--LEVEDVGNMVWKELYQKSFFQDCKMGEYSGDIS-FKMHDLIHDLA 486
LW A F+ N +EDVGN EL +SF QD + SG++ FK+HDL+HDLA
Sbjct: 240 VLWEALDFLPSPNKGKTLEDVGNQFLHELQSRSFLQDFYV---SGNVCVFKLHDLVHDLA 70
Query: 487 QSVMGQECMYLENAN 501
V E L+ N
Sbjct: 69 LYVARDEFQLLKFHN 25
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.138 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,269,872
Number of Sequences: 36976
Number of extensions: 439984
Number of successful extensions: 2735
Number of sequences better than 10.0: 269
Number of HSP's better than 10.0 without gapping: 2426
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2543
length of query: 877
length of database: 9,014,727
effective HSP length: 105
effective length of query: 772
effective length of database: 5,132,247
effective search space: 3962094684
effective search space used: 3962094684
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC148359.9


