

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148349.13 - phase: 0
(348 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
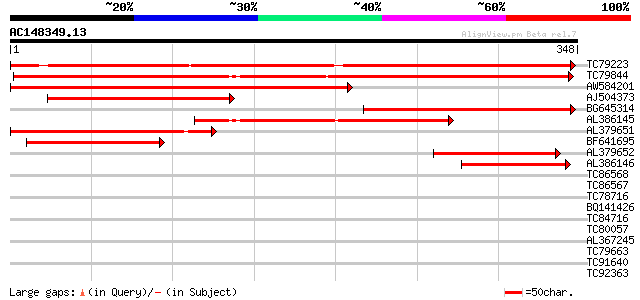
Score E
Sequences producing significant alignments: (bits) Value
TC79223 weakly similar to GP|9759556|dbj|BAB11158.1 steroid sulf... 476 e-135
TC79844 weakly similar to GP|20466686|gb|AAM20660.1 steroid sulf... 410 e-115
AW584201 weakly similar to GP|14595014|emb estrogen sulfotransfe... 358 2e-99
AJ504373 weakly similar to GP|15809903|gb| AT5g07010/MOJ9_18 {Ar... 246 7e-66
BG645314 weakly similar to GP|9759556|dbj| steroid sulfotransfer... 221 4e-58
AL386145 similar to GP|20466686|gb| steroid sulfotransferase-lik... 182 2e-46
AL379651 weakly similar to GP|15809903|gb| AT5g07010/MOJ9_18 {Ar... 166 9e-42
BF641695 weakly similar to PIR|F86407|F864 hypothetical protein ... 144 4e-35
AL379652 weakly similar to GP|15809903|gb| AT5g07010/MOJ9_18 {Ar... 102 2e-22
AL386146 weakly similar to PIR|A84452|A844 probable steroid sulf... 61 6e-10
TC86568 similar to GP|15384991|emb|CAC59824. Xaa-Pro aminopeptid... 30 1.1
TC86567 similar to GP|15384989|emb|CAC59823. Xaa-Pro aminopeptid... 30 1.1
TC78716 weakly similar to PIR|E84652|E84652 hypothetical protein... 29 3.1
BQ141426 29 3.1
TC84716 homologue to GP|17064160|gb|AAL35287.1 WRKY transcriptio... 29 3.1
TC80057 similar to GP|23193174|gb|AAN14410.1 bifunctional lysine... 28 5.2
AL367245 similar to GP|5923674|gb| unknown protein {Arabidopsis ... 28 6.8
TC79663 similar to PIR|F71619|F71619 hypothetical protein PFB023... 28 6.8
TC91640 28 6.8
TC92363 similar to SP|Q9PQH1|IF2_UREPA Translation initiation fa... 27 8.9
>TC79223 weakly similar to GP|9759556|dbj|BAB11158.1 steroid
sulfotransferase-like protein {Arabidopsis thaliana},
partial (47%)
Length = 1208
Score = 476 bits (1225), Expect = e-135
Identities = 230/347 (66%), Positives = 278/347 (79%)
Frame = +2
Query: 1 MASTDLTNMKEDQSRDGQETTSKVNKVSHDYNNLILSLPRENGCETQYMYFFHGFWCPST 60
MAST+LT+ ++Q +D Q + V + NLILSLP+E G QY YFF GFW +
Sbjct: 20 MASTNLTHSTQEQLKDEQ-----LEAVGEECKNLILSLPKEYGFGNQYYYFFQGFWTTPS 184
Query: 61 LIQSVNSFQNNFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPH 120
IQS+ SFQN+F AKDSD+V+AS+PKSGTTWLK L +AI+NR HF+SLEN HPLL NPH
Sbjct: 185 QIQSIISFQNDFQAKDSDVVIASMPKSGTTWLKALTFAIINRHHFSSLEN-HPLLKTNPH 361
Query: 121 ELVPQFEVNLYGDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPF 180
EL+P FE ++Y D P++D+ NM EPRLFGTH+PF SL KS+KESNCKIIYICRNPF
Sbjct: 362 ELIPFFEFHIYVDTVCQFPKVDLLNMIEPRLFGTHIPFTSLAKSIKESNCKIIYICRNPF 541
Query: 181 DTFVSYWIFINKIRLRKSLTELTLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVL 240
DTFVSYW F+NKI L +SL E+ FERYCKGIC GPFWD+MLGYLKESI RP+++L
Sbjct: 542 DTFVSYWNFMNKIELNQSL-----EDDFERYCKGICHVGPFWDHMLGYLKESIARPNKIL 706
Query: 241 FLKYEDLKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTIS 300
FL+YEDLKED+NFH KRIAEF+G PFTQEEE+NGVIENII LCSFE++KE++ N+SG +
Sbjct: 707 FLQYEDLKEDINFHVKRIAEFLGCPFTQEEESNGVIENIINLCSFETLKELDANKSGGLG 886
Query: 301 GDIEKEFYFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSFKVC 347
IEK++YFRK EIGDW NYLS SMVEKLSK++EEKL+GS +SFKVC
Sbjct: 887 WIIEKKYYFRKAEIGDWINYLSPSMVEKLSKIIEEKLSGSGISFKVC 1027
>TC79844 weakly similar to GP|20466686|gb|AAM20660.1 steroid
sulfotransferase-like protein {Arabidopsis thaliana},
partial (69%)
Length = 1379
Score = 410 bits (1055), Expect = e-115
Identities = 202/344 (58%), Positives = 249/344 (71%)
Frame = +1
Query: 3 STDLTNMKEDQSRDGQETTSKVNKVSHDYNNLILSLPRENGCETQYMYFFHGFWCPSTLI 62
S T + + D E+ + ++S + LILSLPRE G T Y+Y F GFWC I
Sbjct: 58 SMSSTKFAKIHTMDIDESIEEQGQLSQENKKLILSLPREKGWRTPYIYLFQGFWCQPAEI 237
Query: 63 QSVNSFQNNFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHEL 122
Q++ +FQ +F AK+SD+ VA++PKSGTTWLK L YAI+NRQ+ NHPLL FNPH+L
Sbjct: 238 QAITTFQKHFQAKESDVFVATVPKSGTTWLKALTYAIMNRQNHFISSKNHPLLSFNPHDL 417
Query: 123 VPQFEVNLYGDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNPFDT 182
VP E +YG D +P D+S + EPRLFGTH+PF SL S+K SNCKI+YICRNPFDT
Sbjct: 418 VPFIEYTVYGKHD-TIP--DLSKIHEPRLFGTHIPFDSLSNSIKGSNCKIVYICRNPFDT 588
Query: 183 FVSYWIFINKIRLRKSLTELTLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFL 242
F+S W F NKI+ SL L LEE+FE YC G+ FGPFW++MLGY ES ERP VLFL
Sbjct: 589 FISSWTFANKIK-PPSLPTLNLEEAFEMYCNGLVGFGPFWNHMLGYWNESKERPKNVLFL 765
Query: 243 KYEDLKEDVNFHTKRIAEFVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGD 302
KYED+KED+ FH K++AEF+ PFT EEE+ GVIENIIKLCSFE MK +E N +GT +
Sbjct: 766 KYEDMKEDLKFHLKKMAEFLDRPFTLEEESEGVIENIIKLCSFEKMKGLEINNTGTFGRN 945
Query: 303 IEKEFYFRKGEIGDWANYLSSSMVEKLSKVMEEKLNGSSLSFKV 346
E +F FRKGE+GDW+NYLS SMVEKLSKV+EEKL GS L+F+V
Sbjct: 946 FENKFLFRKGEVGDWSNYLSPSMVEKLSKVIEEKLGGSDLNFRV 1077
>AW584201 weakly similar to GP|14595014|emb estrogen sulfotransferase {Rattus
norvegicus}, partial (14%)
Length = 669
Score = 358 bits (919), Expect = 2e-99
Identities = 173/211 (81%), Positives = 187/211 (87%), Gaps = 1/211 (0%)
Frame = +2
Query: 1 MASTDLTNMKEDQSRDGQETTSKVNKVSHDYNNLILSLPREN-GCETQYMYFFHGFWCPS 59
MASTDL +MKEDQSRDGQE S+ NKV HDYNNLILSLPRE+ G +QY+YF+H FWCPS
Sbjct: 35 MASTDLKHMKEDQSRDGQEKISEENKVIHDYNNLILSLPREDDGSVSQYLYFYHEFWCPS 214
Query: 60 TLIQSVNSFQNNFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNP 119
T IQS SFQNNFHAKDSDI+ AS+PKSG+TWLKGLAYAIVNRQ+FTSLEN HPLLL NP
Sbjct: 215 TFIQSTISFQNNFHAKDSDIIAASMPKSGSTWLKGLAYAIVNRQYFTSLENKHPLLLSNP 394
Query: 120 HELVPQFEVNLYGDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNP 179
HELVPQFE +LYG KD L Q+DV NMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNP
Sbjct: 395 HELVPQFEASLYGGKDPVLTQVDVPNMTEPRLFGTHMPFPSLPKSVKESNCKIIYICRNP 574
Query: 180 FDTFVSYWIFINKIRLRKSLTELTLEESFER 210
FD FVSYW F NKIRL+KSLTELTLEE+FER
Sbjct: 575 FDIFVSYWTFFNKIRLKKSLTELTLEEAFER 667
>AJ504373 weakly similar to GP|15809903|gb| AT5g07010/MOJ9_18 {Arabidopsis
thaliana}, partial (18%)
Length = 346
Score = 246 bits (629), Expect = 7e-66
Identities = 115/115 (100%), Positives = 115/115 (100%)
Frame = +2
Query: 24 VNKVSHDYNNLILSLPRENGCETQYMYFFHGFWCPSTLIQSVNSFQNNFHAKDSDIVVAS 83
VNKVSHDYNNLILSLPRENGCETQYMYFFHGFWCPSTLIQSVNSFQNNFHAKDSDIVVAS
Sbjct: 2 VNKVSHDYNNLILSLPRENGCETQYMYFFHGFWCPSTLIQSVNSFQNNFHAKDSDIVVAS 181
Query: 84 IPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQFEVNLYGDKDGPL 138
IPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQFEVNLYGDKDGPL
Sbjct: 182 IPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPHELVPQFEVNLYGDKDGPL 346
>BG645314 weakly similar to GP|9759556|dbj| steroid sulfotransferase-like
protein {Arabidopsis thaliana}, partial (20%)
Length = 671
Score = 221 bits (562), Expect = 4e-58
Identities = 103/130 (79%), Positives = 117/130 (89%)
Frame = -1
Query: 218 FGPFWDNMLGYLKESIERPDRVLFLKYEDLKEDVNFHTKRIAEFVGIPFTQEEENNGVIE 277
FGPFW+NMLGY KESIE P RVLFLKYEDLKEDVNF+ KRIAEFVGIPFTQEEENNGVIE
Sbjct: 671 FGPFWENMLGYFKESIESPYRVLFLKYEDLKEDVNFNIKRIAEFVGIPFTQEEENNGVIE 492
Query: 278 NIIKLCSFESMKEIEGNQSGTISGDIEKEFYFRKGEIGDWANYLSSSMVEKLSKVMEEKL 337
NIIKLCSFESMKE GN SGT++ ++++EF+FRKGEIGDW NY S SM+E+LSK+M+EK
Sbjct: 491 NIIKLCSFESMKESNGNNSGTVTVNVDREFFFRKGEIGDWVNYFSPSMIERLSKIMQEKF 312
Query: 338 NGSSLSFKVC 347
+GSSLSFK C
Sbjct: 311 SGSSLSFKGC 282
>AL386145 similar to GP|20466686|gb| steroid sulfotransferase-like protein
{Arabidopsis thaliana}, partial (15%)
Length = 467
Score = 182 bits (461), Expect = 2e-46
Identities = 86/159 (54%), Positives = 122/159 (76%)
Frame = +3
Query: 114 LLLFNPHELVPQFEVNLYGDKDGPLPQIDVSNMTEPRLFGTHMPFPSLPKSVKESNCKII 173
LL NPH LVP E++LY +KD LP D+++ + PRLF TH+P+ SLPKS+K+S CK++
Sbjct: 3 LLTTNPHVLVPFLELSLYIEKD-ILP--DINSFSAPRLFSTHVPYKSLPKSIKDSTCKVV 173
Query: 174 YICRNPFDTFVSYWIFINKIRLRKSLTELTLEESFERYCKGICLFGPFWDNMLGYLKESI 233
Y+CR+P DTF S W F NKIR + T L LEESFE++ +G+ LFGPFW+++LGY KES+
Sbjct: 174 YLCRDPKDTFASMWHFTNKIRPQNRET-LQLEESFEKFSRGVSLFGPFWEHLLGYWKESL 350
Query: 234 ERPDRVLFLKYEDLKEDVNFHTKRIAEFVGIPFTQEEEN 272
ER ++V+FL+YE++K F+ K +AEF+G PF++EEE+
Sbjct: 351 ERQEKVMFLRYEEMKMKPCFYLKEVAEFLGCPFSKEEES 467
>AL379651 weakly similar to GP|15809903|gb| AT5g07010/MOJ9_18 {Arabidopsis
thaliana}, partial (14%)
Length = 400
Score = 166 bits (421), Expect = 9e-42
Identities = 83/127 (65%), Positives = 94/127 (73%)
Frame = +2
Query: 1 MASTDLTNMKEDQSRDGQETTSKVNKVSHDYNNLILSLPRENGCETQYMYFFHGFWCPST 60
MASTDLTNMKEDQSRDGQET S+ NKV HDYNNLILSLP+ENG +F FWCPS
Sbjct: 20 MASTDLTNMKEDQSRDGQETISEENKVIHDYNNLILSLPKENGWGLSQNVYFQCFWCPSN 199
Query: 61 LIQSVNSFQNNFHAKDSDIVVASIPKSGTTWLKGLAYAIVNRQHFTSLENNHPLLLFNPH 120
+IQ + SFQNNF AK +DIVVAS+PKSGTTWL+ + IVNR ++ NHPLL H
Sbjct: 200 MIQPIISFQNNFQAKHNDIVVASLPKSGTTWLRAFTFTIVNRNQYSF--ENHPLLKSIIH 373
Query: 121 ELVPQFE 127
ELVP E
Sbjct: 374 ELVPSIE 394
>BF641695 weakly similar to PIR|F86407|F864 hypothetical protein AAF98439.1
[imported] - Arabidopsis thaliana, partial (10%)
Length = 656
Score = 144 bits (364), Expect = 4e-35
Identities = 66/85 (77%), Positives = 74/85 (86%)
Frame = +1
Query: 11 EDQSRDGQETTSKVNKVSHDYNNLILSLPRENGCETQYMYFFHGFWCPSTLIQSVNSFQN 70
++Q+ T + NKV HDYNNLILSLPRENGCET+YMYFFHGFWCPS LIQSVN+FQN
Sbjct: 400 QEQNPPMASTYNTENKVIHDYNNLILSLPRENGCETEYMYFFHGFWCPSYLIQSVNTFQN 579
Query: 71 NFHAKDSDIVVASIPKSGTTWLKGL 95
FHAKD+DIVVAS+PKSGTTWLKGL
Sbjct: 580 KFHAKDNDIVVASMPKSGTTWLKGL 654
>AL379652 weakly similar to GP|15809903|gb| AT5g07010/MOJ9_18 {Arabidopsis
thaliana}, partial (9%)
Length = 488
Score = 102 bits (254), Expect = 2e-22
Identities = 46/78 (58%), Positives = 62/78 (78%)
Frame = +1
Query: 261 FVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDIEKEFYFRKGEIGDWANY 320
F+G FTQEEE+ V+ENII LC FE+MKE+E N+ G++ DIE +F+FRK ++ DW NY
Sbjct: 4 FLGYSFTQEEESKKVVENIINLCCFETMKELEVNKFGSVRSDIENKFFFRKAKVEDWKNY 183
Query: 321 LSSSMVEKLSKVMEEKLN 338
LS SM EKLSK+++EKL+
Sbjct: 184 LSPSMEEKLSKIVDEKLS 237
>AL386146 weakly similar to PIR|A84452|A844 probable steroid sulfotransferase
[imported] - Arabidopsis thaliana, partial (12%)
Length = 494
Score = 61.2 bits (147), Expect = 6e-10
Identities = 26/67 (38%), Positives = 42/67 (61%)
Frame = +1
Query: 278 NIIKLCSFESMKEIEGNQSGTISGDIEKEFYFRKGEIGDWANYLSSSMVEKLSKVMEEKL 337
+I+ LCSFE + +E N+ G + E + +FR G++GDW LS M+E+L+ V+E+ L
Sbjct: 1 DILNLCSFEKLSNLEVNKFGKLPSGEENKAFFRSGKVGDWKTLLSIEMIERLNTVIEQNL 180
Query: 338 NGSSLSF 344
+SF
Sbjct: 181 GKHGMSF 201
>TC86568 similar to GP|15384991|emb|CAC59824. Xaa-Pro aminopeptidase 2
{Lycopersicon esculentum}, partial (27%)
Length = 823
Score = 30.4 bits (67), Expect = 1.1
Identities = 15/54 (27%), Positives = 26/54 (47%), Gaps = 8/54 (14%)
Frame = +3
Query: 48 YMYFFHGFWCP--------STLIQSVNSFQNNFHAKDSDIVVASIPKSGTTWLK 93
Y+ F H W P + L ++ N++H+K DI+ + ++G WLK
Sbjct: 381 YLSFEHITWAPYQTKLIDLNLLNPEEKNWLNSYHSKCRDILAPHLDEAGNAWLK 542
>TC86567 similar to GP|15384989|emb|CAC59823. Xaa-Pro aminopeptidase 1
{Lycopersicon esculentum}, partial (91%)
Length = 2223
Score = 30.4 bits (67), Expect = 1.1
Identities = 15/54 (27%), Positives = 26/54 (47%), Gaps = 8/54 (14%)
Frame = +1
Query: 48 YMYFFHGFWCP--------STLIQSVNSFQNNFHAKDSDIVVASIPKSGTTWLK 93
Y+ F H W P + L ++ N++H+K DI+ + ++G WLK
Sbjct: 1825 YLSFEHITWAPYQTKLIDLNLLNPEEKNWLNSYHSKCRDILAPHLDEAGNAWLK 1986
>TC78716 weakly similar to PIR|E84652|E84652 hypothetical protein At2g25770
[imported] - Arabidopsis thaliana, partial (22%)
Length = 789
Score = 28.9 bits (63), Expect = 3.1
Identities = 9/22 (40%), Positives = 16/22 (71%)
Frame = +2
Query: 29 HDYNNLILSLPRENGCETQYMY 50
H Y + + LP++NGCE +++Y
Sbjct: 326 HSYVSTVRVLPKDNGCEIEWIY 391
>BQ141426
Length = 471
Score = 28.9 bits (63), Expect = 3.1
Identities = 16/67 (23%), Positives = 32/67 (46%), Gaps = 5/67 (7%)
Frame = +1
Query: 225 MLGYLKESIERPDRVLFLKYEDLKEDVN-----FHTKRIAEFVGIPFTQEEENNGVIENI 279
M+ Y E + ++L + +LK VN FH K + + +P ++ + + +
Sbjct: 145 MMSYYLEVFQNSQKLLMSLFMNLKSFVNLISILFHLKPSLQVL*LPLKSLNKSKKLKQLL 324
Query: 280 IKLCSFE 286
IK+C F+
Sbjct: 325 IKICKFQ 345
>TC84716 homologue to GP|17064160|gb|AAL35287.1 WRKY transcription factor 29
{Arabidopsis thaliana}, partial (21%)
Length = 524
Score = 28.9 bits (63), Expect = 3.1
Identities = 16/67 (23%), Positives = 32/67 (46%), Gaps = 5/67 (7%)
Frame = +3
Query: 225 MLGYLKESIERPDRVLFLKYEDLKEDVN-----FHTKRIAEFVGIPFTQEEENNGVIENI 279
M+ Y E + ++L + +LK VN FH K + + +P ++ + + +
Sbjct: 69 MMSYYLEVFQNSQKLLMSLFMNLKSFVNLISILFHLKPSLQVL*LPLKSLNKSKKLKQLL 248
Query: 280 IKLCSFE 286
IK+C F+
Sbjct: 249 IKICKFQ 269
>TC80057 similar to GP|23193174|gb|AAN14410.1 bifunctional
lysine-ketoglutarate reductase/saccharopine
dehydrogenase {Gossypium hirsutum}, partial (38%)
Length = 1405
Score = 28.1 bits (61), Expect = 5.2
Identities = 18/64 (28%), Positives = 28/64 (43%), Gaps = 11/64 (17%)
Frame = -2
Query: 21 TSKVNKVSHDYNNLILSLPRE-----------NGCETQYMYFFHGFWCPSTLIQSVNSFQ 69
++K +SH NL+++ R NGC + YFF +W ++ V F
Sbjct: 363 SNKWTSISHQILNLLIAQQRRKPFLPIHAINNNGCV--WCYFFMEYWIVMFRMRIVVYFV 190
Query: 70 NNFH 73
NFH
Sbjct: 189 KNFH 178
>AL367245 similar to GP|5923674|gb| unknown protein {Arabidopsis thaliana},
partial (14%)
Length = 391
Score = 27.7 bits (60), Expect = 6.8
Identities = 24/112 (21%), Positives = 48/112 (42%)
Frame = +2
Query: 201 ELTLEESFERYCKGICLFGPFWDNMLGYLKESIERPDRVLFLKYEDLKEDVNFHTKRIAE 260
E + SFE + L + + Y+ ES+ + + K+E L++ VN +
Sbjct: 35 EADADSSFELFRNSDDLADEYNYDYDDYVDESLWGDEEWIEGKHEKLEDYVNVDS----H 202
Query: 261 FVGIPFTQEEENNGVIENIIKLCSFESMKEIEGNQSGTISGDIEKEFYFRKG 312
+ P + +N+GV+E ++ + F + + + GDI+ Y G
Sbjct: 203 ILSTPVIADIDNDGVMEMVVAVSYFFDQEYYDNQEHMKELGDIDIGKYVAGG 358
>TC79663 similar to PIR|F71619|F71619 hypothetical protein PFB0235w -
malaria parasite (Plasmodium falciparum), partial (4%)
Length = 1166
Score = 27.7 bits (60), Expect = 6.8
Identities = 13/37 (35%), Positives = 20/37 (53%), Gaps = 9/37 (24%)
Frame = -2
Query: 26 KVSHDYNNLILS---------LPRENGCETQYMYFFH 53
K SH+ NN+++S +P+ NG + YFFH
Sbjct: 232 KHSHNLNNILMSHFFTKRHRIIPKTNGNRIRVTYFFH 122
>TC91640
Length = 541
Score = 27.7 bits (60), Expect = 6.8
Identities = 13/33 (39%), Positives = 16/33 (48%)
Frame = +3
Query: 255 TKRIAEFVGIPFTQEEENNGVIENIIKLCSFES 287
T + F+GIP E +I IKL SF S
Sbjct: 150 TSELCTFMGIPHKSRSEIASIISKFIKLYSFRS 248
>TC92363 similar to SP|Q9PQH1|IF2_UREPA Translation initiation factor IF-2.
[Ureaplasma urealyticum biotype 1] {Ureaplasma parvum},
partial (2%)
Length = 849
Score = 27.3 bits (59), Expect = 8.9
Identities = 16/40 (40%), Positives = 22/40 (55%)
Frame = -1
Query: 172 IIYICRNPFDTFVSYWIFINKIRLRKSLTELTLEESFERY 211
IIY CR F ++Y I K++ K+ L LEE E+Y
Sbjct: 501 IIYDCREFFPCTITYEANIEKLKEEKN---LKLEEDCEKY 391
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,267,140
Number of Sequences: 36976
Number of extensions: 197126
Number of successful extensions: 1016
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 1003
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1008
length of query: 348
length of database: 9,014,727
effective HSP length: 97
effective length of query: 251
effective length of database: 5,428,055
effective search space: 1362441805
effective search space used: 1362441805
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148349.13


