

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148347.4 + phase: 1 /pseudo
(776 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
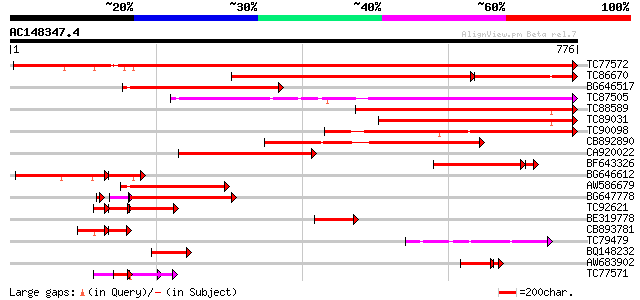
Score E
Sequences producing significant alignments: (bits) Value
TC77572 similar to GP|9558523|dbj|BAB03441.1 ESTs AU078742(C1188... 1281 0.0
TC86670 weakly similar to GP|9758141|dbj|BAB08633.1 disease resi... 467 0.0
BG646517 380 e-106
TC87505 weakly similar to PIR|T05981|T05981 hypothetical protein... 327 8e-90
TC88589 weakly similar to GP|9758140|dbj|BAB08632.1 disease resi... 308 5e-84
TC89031 weakly similar to GP|9758141|dbj|BAB08633.1 disease resi... 303 2e-82
TC90098 weakly similar to GP|9758140|dbj|BAB08632.1 disease resi... 295 3e-80
CB892890 weakly similar to GP|9758141|dbj| disease resistance pr... 293 2e-79
CA920022 similar to PIR|T03031|T030 NBS-LRR type resistance prot... 268 6e-72
BF643326 weakly similar to GP|11994216|db disease resistance com... 204 3e-56
BG646612 weakly similar to GP|9758140|dbj| disease resistance pr... 169 3e-53
AW586679 similar to GP|9758140|dbj| disease resistance protein-l... 149 5e-36
BG647778 weakly similar to PIR|T48468|T484 disease resistance-li... 120 2e-29
TC92621 similar to GP|7107252|gb|AAF36340.1| unknown {Pennisetum... 97 2e-27
BE319778 119 4e-27
CB893781 homologue to PIR|C64504|C645 hypothetical protein MJ163... 68 6e-23
TC79479 similar to GP|12056928|gb|AAG48132.1 putative resistance... 88 1e-17
BQ148232 similar to GP|9758141|dbj| disease resistance protein-l... 78 1e-14
AW683902 weakly similar to PIR|T48468|T484 disease resistance-li... 67 4e-13
TC77571 homologue to GP|20804846|dbj|BAB92528. putative rust res... 57 1e-11
>TC77572 similar to GP|9558523|dbj|BAB03441.1 ESTs AU078742(C11888)
C26225(C11888) correspond to a region of the predicted
gene.~Similar to Oryza, partial (5%)
Length = 2535
Score = 1281 bits (3315), Expect = 0.0
Identities = 676/805 (83%), Positives = 703/805 (86%), Gaps = 34/805 (4%)
Frame = +3
Query: 6 TYYYPYNKI--**CLEQNI*NS*NSTKEPKFKAIV*VNLKGFISRCTRYKALQ*TLGSTQ 63
T+YYPYNKI **CLEQNI NS STKEPKFKAI+ VNLKGFISRCTRYKA+Q*+ GS Q
Sbjct: 51 THYYPYNKISI**CLEQNISNSSKSTKEPKFKAIIEVNLKGFISRCTRYKAIQ*SFGSPQ 230
Query: 64 RRNK*FDRRK-----CMQIFF*E*FLR--C**EPISYCK*CRRDTLQGQRNS*ASQPR-- 114
RRNK* DRRK CM +FF*E*FL C* EPISYCK*C RD L QRNS*ASQP
Sbjct: 231 RRNK*SDRRK*CRRKCMYMFF*E*FLWLCC*GEPISYCK*C*RDAL*DQRNS*ASQP*IQ 410
Query: 115 --------------KSKIHCWVGYTI*QVENGASEGVVAPHLC*LV*EDWGKPLW----- 155
KSKIHCWVGYTI*QVENGASEG H C DW + +
Sbjct: 411 *RWTTFQASFQCS*KSKIHCWVGYTI*QVENGASEGW*L-HTC----VDWFRRIGENHSG 575
Query: 156 LQSFVGMKKLMEN----IIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLL 211
++ +G KKLM N + K P + VERIHQHCGY VPEFQSDEDAVN+LGLL
Sbjct: 576 YKALLG*KKLMVNSRKTLYLSPSQKHPC*RLFVERIHQHCGYPVPEFQSDEDAVNRLGLL 755
Query: 212 LKKVEGSPLLLVLDDVWPSSESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHED 271
LKKVEGSPLLLVLDDVWP SE LVEK+QFQ++ FKILVTSRVAFPRFSTTCILKPLAHE+
Sbjct: 756 LKKVEGSPLLLVLDDVWPISEPLVEKIQFQISDFKILVTSRVAFPRFSTTCILKPLAHEE 935
Query: 272 AVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKEL 331
AVTLFHHYAQMEKNSSDII+KNLVEKVVRSC GLPLTIKVIATSLKNRP D+W KI KEL
Sbjct: 936 AVTLFHHYAQMEKNSSDIINKNLVEKVVRSCQGLPLTIKVIATSLKNRPRDMWRKIGKEL 1115
Query: 332 SQGHSILDSNTELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYK 391
SQGHSILDSNT+LLTRLQKIFDVLEDNP IMECFMD+ALFPEDHRIPVAALVDMWAELY+
Sbjct: 1116SQGHSILDSNTDLLTRLQKIFDVLEDNPTIMECFMDIALFPEDHRIPVAALVDMWAELYR 1295
Query: 392 LDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILHDILRELGIYQSTKE 451
LDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILHDILRELGIYQS KE
Sbjct: 1296LDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILHDILRELGIYQSAKE 1475
Query: 452 PFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCA 511
PFEQRKRLIID+N NKSGLAEKQQGLMT I SKFMRLCVK+NPQQL ARILSVS DETCA
Sbjct: 1476PFEQRKRLIIDINKNKSGLAEKQQGLMTCILSKFMRLCVKRNPQQLTARILSVSADETCA 1655
Query: 512 LDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQ 571
DWS M+PAQVEVLILN+HTKQYSLPEWIAKMSKLRVLIITNY FHPSKLNNIELLGSLQ
Sbjct: 1656FDWSQMEPAQVEVLILNLHTKQYSLPEWIAKMSKLRVLIITNYDFHPSKLNNIELLGSLQ 1835
Query: 572 NLERIRLERISVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDL 631
NLERIRLERI VPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDL
Sbjct: 1836NLERIRLERIYVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDL 2015
Query: 632 VVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLN 691
VVL GICDI LKKL VTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLN
Sbjct: 2016VVLPIGICDIFLLKKLRVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLN 2195
Query: 692 LKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASIELPFSVVNLQNLKTITCDEETAAT 751
LKHLDISNCISLSSLPEEFGNLCNLKNLDMA+CASIELPFSVVNLQNLKTITCDEETAAT
Sbjct: 2196LKHLDISNCISLSSLPEEFGNLCNLKNLDMASCASIELPFSVVNLQNLKTITCDEETAAT 2375
Query: 752 WEDFQHMLHNMKIEVLHVDVNLNWL 776
WEDFQHML NMKIEVLHVDVNLNWL
Sbjct: 2376WEDFQHMLPNMKIEVLHVDVNLNWL 2450
Score = 31.2 bits (69), Expect = 1.6
Identities = 16/44 (36%), Positives = 22/44 (49%)
Frame = -3
Query: 668 LELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFG 711
L L++S L +P S GKL L ++S C +S P E G
Sbjct: 2299 LAQLAMSRFFRLHKLPNSSGKLERLMQFEMSRCFRFNSFPMEVG 2168
>TC86670 weakly similar to GP|9758141|dbj|BAB08633.1 disease resistance
protein-like {Arabidopsis thaliana}, partial (7%)
Length = 2417
Score = 467 bits (1201), Expect(2) = 0.0
Identities = 237/336 (70%), Positives = 273/336 (80%), Gaps = 2/336 (0%)
Frame = +3
Query: 304 GLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILDSNTELLTRLQKIFDVLEDNPKIME 363
GLPL IKVIATS + RP++LW KIVKELS+G SILDSNTELL RLQKI DVLEDN E
Sbjct: 6 GLPLAIKVIATSFRYRPYELWEKIVKELSRGRSILDSNTELLIRLQKILDVLEDNAISKE 185
Query: 364 CFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASD 423
CFMDLALFPED RIPVAAL+DMWAELY LDD+G +AM+IINKL MNLANV+I RKNASD
Sbjct: 186 CFMDLALFPEDQRIPVAALIDMWAELYGLDDDGKEAMDIINKLDSMNLANVLIARKNASD 365
Query: 424 TDNNNYNNHFIILHDILRELGIYQSTKEPFEQRKRLIIDMNNNK--SGLAEKQQGLMTRI 481
T+N YNNHFI+LHD+LRELG YQ+T+EP EQRKR +I+ N +K L EKQQG M I
Sbjct: 366 TENYYYNNHFIVLHDLLRELGNYQNTQEPIEQRKRQLINTNESKCDQRLREKQQGTMAHI 545
Query: 482 FSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIA 541
SK + K PQ++ AR LS+S DETCA DWS +QPA EVLILN+ TKQY+ PE +
Sbjct: 546 LSKLIGWFDKPKPQKVPARTLSISIDETCASDWSQVQPALAEVLILNLQTKQYTFPELME 725
Query: 542 KMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISVPSFGTLKNLKKLSLYMCN 601
KM+KL+ LI+ N+G PS+LNN+ELL SL NL+RIRLERISVPSFGTLKNLKKLSLYMCN
Sbjct: 726 KMNKLKALIVINHGLRPSELNNLELLSSLSNLKRIRLERISVPSFGTLKNLKKLSLYMCN 905
Query: 602 TILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTG 637
T LAFEKGSILISD FPNLE+L++DYCKD+ L G
Sbjct: 906 TRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNG 1013
Score = 226 bits (577), Expect(2) = 0.0
Identities = 110/140 (78%), Positives = 125/140 (88%)
Frame = +1
Query: 637 GICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLD 696
G+CDIISLKKL++TNCHKLS LPQ+IGKLENLELLSL SCTDL +P SIG+L NL+ LD
Sbjct: 1012 GVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLD 1191
Query: 697 ISNCISLSSLPEEFGNLCNLKNLDMATCASIELPFSVVNLQNLKTITCDEETAATWEDFQ 756
ISNCISLSSLPE+FGNLCNL+NLDM +CAS ELPFSVVNLQNLK +TCDEETAA+WE FQ
Sbjct: 1192 ISNCISLSSLPEDFGNLCNLRNLDMTSCASCELPFSVVNLQNLK-VTCDEETAASWESFQ 1368
Query: 757 HMLHNMKIEVLHVDVNLNWL 776
M+ N+ IEV HV+VNLNWL
Sbjct: 1369 SMISNLTIEVPHVEVNLNWL 1428
Score = 29.6 bits (65), Expect = 4.6
Identities = 17/45 (37%), Positives = 24/45 (52%)
Frame = -1
Query: 668 LELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGN 712
L L +S L +P S GKL L L++S+ + SLP E G+
Sbjct: 1280 LAQLVMSRFRRLHRLPKSSGKLERLMQLEMSSSLRFESLPIESGS 1146
>BG646517
Length = 668
Score = 380 bits (977), Expect = e-106
Identities = 193/220 (87%), Positives = 202/220 (91%)
Frame = +2
Query: 155 WLQSFVGMKKLMENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLKK 214
W Q G K ENIIFVTFSKTPMLKTIVERI +HCGY VPEFQSDEDAVNKL LLLKK
Sbjct: 8 WDQEVNG--KFKENIIFVTFSKTPMLKTIVERIIEHCGYPVPEFQSDEDAVNKLELLLKK 181
Query: 215 VEGSPLLLVLDDVWPSSESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVT 274
VEGSPLLLVLDDVWP+SESLV+KLQF+++ FKILVTSRV+FPRF TTCILKPLAHEDAVT
Sbjct: 182 VEGSPLLLVLDDVWPTSESLVKKLQFEISDFKILVTSRVSFPRFRTTCILKPLAHEDAVT 361
Query: 275 LFHHYAQMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQG 334
LFHHYAQMEKNSSDIIDKNLVEKVVRSC GLPLTIKVIATSL+NRP DLW KIV ELSQG
Sbjct: 362 LFHHYAQMEKNSSDIIDKNLVEKVVRSCKGLPLTIKVIATSLRNRPDDLWRKIVMELSQG 541
Query: 335 HSILDSNTELLTRLQKIFDVLEDNPKIMECFMDLALFPED 374
HSILDSNTELLTRLQKIFDVLEDNP IMECFMD+ALFPED
Sbjct: 542 HSILDSNTELLTRLQKIFDVLEDNPTIMECFMDIALFPED 661
>TC87505 weakly similar to PIR|T05981|T05981 hypothetical protein F17M5.60 -
Arabidopsis thaliana, partial (45%)
Length = 2140
Score = 327 bits (839), Expect = 8e-90
Identities = 191/565 (33%), Positives = 324/565 (56%), Gaps = 8/565 (1%)
Frame = +3
Query: 220 LLLVLDDVWPSSESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHY 279
+L++LDDVW S S++E+L F+M K +V SR FP F+ T ++ L +DA++LF H+
Sbjct: 396 ILVILDDVW--SPSVLEQLVFRMPNCKFIVVSRFIFPIFNATYKVELLDKDDALSLFCHH 569
Query: 280 AQMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILD 339
A +K+ ++NLV++VV C LPL +KVI SL+++ W + LSQG SI +
Sbjct: 570 AFGQKSIPFAANQNLVKQVVAECGNLPLALKVIGASLRDQNEMFWLSVKTRLSQGLSIDE 749
Query: 340 S-NTELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQ 398
S L+ R+ + L + KI ECF+DL FPED +IP+ L++MW E++ + + +
Sbjct: 750 SYERNLIDRMAISTNYLPE--KIKECFLDLCSFPEDKKIPLEVLINMWVEIHDIHET--E 917
Query: 399 AMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHF---IILHDILRELGIYQSTKEPFEQ 455
A I+ +L NL ++ + Y++ F + HDILR+L + S + Q
Sbjct: 918 AYAIVVELSNKNLLTLVEEARAGG-----MYSSCFEISVTQHDILRDLALNLSNRGNINQ 1082
Query: 456 RKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWS 515
R+RL++ + L ++ ++ Q A+I+S+ T E DW
Sbjct: 1083RRRLVMPKREDNGQLPKEW---------------LRYADQPFEAQIVSIHTGEMRKSDWC 1217
Query: 516 HMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLER 575
+++ + EVLI+N + +Y LP +I +M KLR L++ N+ + L+NI + +L NL
Sbjct: 1218NLEFPKAEVLIINFTSSEYFLPPFINRMPKLRALMVINHSTSYACLHNISVFKNLTNLRS 1397
Query: 576 IRLERISVPSFG--TLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVV 633
+ E++S+P +++L+KL + +C + E I+D FPN+ EL +D+C+D+
Sbjct: 1398LWFEKVSIPHLSGIVMESLRKLFIVLCKINNSLEGKDSNIADIFPNISELTLDHCEDVTE 1577
Query: 634 LQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLK 693
L + IC I SL+ L++TNCH L+ LP ++G L LE+L L +C +L +P SI + LK
Sbjct: 1578LPSSICRIQSLQNLSLTNCHSLTRLPIELGSLRYLEILRLYACPNLRTLPPSICGMTRLK 1757
Query: 694 HLDISNCISLSSLPEEFGNLCNLKNLDMATCASI-ELPFSVVNLQNLKTI-TCDEETAAT 751
++DI+ C+ L+S P+ G L NL+ +DM C I +P S ++L ++ CD+E +
Sbjct: 1758YIDIAQCVYLASFPDAIGKLVNLEKIDMRECPMITNIPKSALSLNFFCSL*ICDDEVSWM 1937
Query: 752 WEDFQHMLHNMKIEVLHVDVNLNWL 776
W++ Q + N+ I+V+ ++ +L+WL
Sbjct: 1938WKEVQKVKLNVDIQVVEIEYDLDWL 2012
>TC88589 weakly similar to GP|9758140|dbj|BAB08632.1 disease resistance
protein-like {Arabidopsis thaliana}, partial (12%)
Length = 1137
Score = 308 bits (789), Expect = 5e-84
Identities = 164/307 (53%), Positives = 215/307 (69%), Gaps = 4/307 (1%)
Frame = +1
Query: 474 QQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQ 533
QQG+++ +S + VKQ ++AARIL +STDE + DW MQP + EVL+LN+ + Q
Sbjct: 25 QQGIISNWYSFITGMLVKQKQLKVAARILCISTDEIFSSDWCDMQPDKAEVLVLNLRSDQ 204
Query: 534 YSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISVPSFGTLKNLK 593
YSLP++ KM KL+VLI+TNYGF S+L ELLGSL NL+RIRLE++SVP LKNL+
Sbjct: 205 YSLPDFTKKMRKLKVLIVTNYGFSRSELTKFELLGSLSNLKRIRLEKVSVPCLCILKNLR 384
Query: 594 KLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCH 653
KLSL+MC+T AFE SI ISDA PNL EL+IDYC DL+ L C I +LKKL++TNCH
Sbjct: 385 KLSLHMCSTNNAFESCSIQISDAMPNLVELSIDYCNDLIKLPGEFCKITTLKKLSITNCH 564
Query: 654 KLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNL 713
K S++PQDIGKL NLE+L L SC+DL+ IP S+ L L+ LDIS+C++L LP GNL
Sbjct: 565 KFSAMPQDIGKLVNLEVLRLCSCSDLKEIPESVADLNKLRCLDISDCVTLHILPNNIGNL 744
Query: 714 CNLKNLDMATCASI-ELPFSVVNLQNLK---TITCDEETAATWEDFQHMLHNMKIEVLHV 769
L+ L M C+++ ELP SV+N NLK + CDEE +A WE + + +KI + V
Sbjct: 745 QKLEKLYMKGCSNLSELPDSVINFGNLKHEMQVICDEEGSALWEHLSN-IPKLKIYMPKV 921
Query: 770 DVNLNWL 776
+ NL WL
Sbjct: 922 EHNLIWL 942
>TC89031 weakly similar to GP|9758141|dbj|BAB08633.1 disease resistance
protein-like {Arabidopsis thaliana}, partial (5%)
Length = 1294
Score = 303 bits (775), Expect = 2e-82
Identities = 156/275 (56%), Positives = 204/275 (73%), Gaps = 4/275 (1%)
Frame = +3
Query: 506 TDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIE 565
TDE+ + DW MQP +VEVL+LN+ + QYSLP++ KMSKL+VLI+TNYGFH S+L E
Sbjct: 120 TDESFSSDWCDMQPDEVEVLVLNLQSDQYSLPDFTDKMSKLKVLIVTNYGFHRSELIKFE 299
Query: 566 LLGSLQNLERIRLERISVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNI 625
LLG L NL+RIRLE++SVP LKNL+KLSL+MCNT AFE SI ISDA PNL EL+I
Sbjct: 300 LLGFLSNLKRIRLEKVSVPCLSILKNLQKLSLHMCNTRDAFENYSIQISDAMPNLVELSI 479
Query: 626 DYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTS 685
DYC DL+ L G +I +LKK+++TNCHKLS++PQDI KLENLE+L L SC+DL I S
Sbjct: 480 DYCNDLIKLPDGFSNITTLKKISITNCHKLSAIPQDIEKLENLEVLRLCSCSDLVEISES 659
Query: 686 IGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASI-ELPFSVVNLQNLK---T 741
+ L L+ DIS+C+SLS LP + G+L L+ M C+++ ELP+SV+NL N+K
Sbjct: 660 VSGLNKLRCFDISDCVSLSKLPNDIGDLKKLEKFYMKGCSNLSELPYSVINLGNVKHEIH 839
Query: 742 ITCDEETAATWEDFQHMLHNMKIEVLHVDVNLNWL 776
+ CDEE AA WE F + + N+KI++ V++NLNWL
Sbjct: 840 VICDEEGAALWEHFPN-IPNLKIDMPKVEINLNWL 941
>TC90098 weakly similar to GP|9758140|dbj|BAB08632.1 disease resistance
protein-like {Arabidopsis thaliana}, partial (16%)
Length = 1128
Score = 295 bits (756), Expect = 3e-80
Identities = 165/351 (47%), Positives = 226/351 (64%), Gaps = 6/351 (1%)
Frame = +1
Query: 432 HFIILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVK 491
H++ H +LR+L I +++E ++R RLIID + N
Sbjct: 1 HYVTQHGLLRDLAIRDNSQEAEDKRNRLIIDTSANN-----------------LPSWWTS 129
Query: 492 QNPQQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLII 551
+N + ARILS+STDE W ++QP +VEVL+LN+ K+++LP ++ KM KL+ LII
Sbjct: 130 ENEYHIKARILSISTDEIFTSKWCNLQPTEVEVLVLNLREKKFALPMFMKKMKKLKALII 309
Query: 552 TNYGFHPSKLNNIELLGSLQNLERIRLERISVPSFG----TLKNLKKLSLYMCNTILAFE 607
TNY F P++L N E+L L NL+RIRLE++SVP G LKNL+K S +MCN AFE
Sbjct: 310 TNYDFFPAELENFEVLDHLSNLKRIRLEKVSVPFLGKTVLQLKNLQKCSFFMCNVNKAFE 489
Query: 608 KGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLEN 667
+I S+ PNL E+N DYC D+V L I +I+SLKKL++TNCHKL +L + IGKL N
Sbjct: 490 NCTIEDSEMLPNLMEMNFDYC-DMVELPNVISNIVSLKKLSITNCHKLCALHEGIGKLVN 666
Query: 668 LELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASI 727
LE L LSSC+ L +P SI L LK L+IS CISLS LPE G L L+ L+M C++I
Sbjct: 667 LESLRLSSCSGLLELPNSIRNLHVLKFLNISGCISLSQLPENIGELEKLEKLNMRGCSNI 846
Query: 728 -ELPFSVVNLQNLKTITC-DEETAATWEDFQHMLHNMKIEVLHVDVNLNWL 776
ELP SV+ L+ LK + C DEETA WE ++++L ++K+EV+ D NLN+L
Sbjct: 847 SELPSSVMELEGLKHVVCGDEETAEKWEPYKNILGDLKVEVVQEDFNLNFL 999
>CB892890 weakly similar to GP|9758141|dbj| disease resistance protein-like
{Arabidopsis thaliana}, partial (4%)
Length = 852
Score = 293 bits (750), Expect = 2e-79
Identities = 155/305 (50%), Positives = 208/305 (67%), Gaps = 3/305 (0%)
Frame = +2
Query: 349 QKIFD-VLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLG 407
QK+ + L+DN + ECFMDL LF ED +IPVAAL+DMW ELY LDD+ I+ M I+ KL
Sbjct: 2 QKVLENALQDNHIVKECFMDLGLFFEDKKIPVAALIDMWTELYDLDDDNIKGMNIVRKLA 181
Query: 408 IMNLANVIIPRKNASDTDNNNYNNHFIILHDILRELGIYQSTKEPFEQRKRLIIDMNNNK 467
+L +++ R+ + D+ YN+HF+ HD+L+E+ I+Q+ +EPFE RKRLI D+N N
Sbjct: 182 NWHLVKLVVSREVTTHVDHY-YNHHFLTQHDLLKEISIHQARQEPFELRKRLIFDVNENS 358
Query: 468 SGLAEKQQGLMTRIFSKFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPA-QVEVLI 526
+QN Q AR LS+S D+ DWS+++ QVEVLI
Sbjct: 359 WD---------------------QQNQQNTIARTLSISPDKILTSDWSNVEKIKQVEVLI 475
Query: 527 LNIHTKQYSLPEWIAKMSKLRVLIITNY-GFHPSKLNNIELLGSLQNLERIRLERISVPS 585
LN+HT++Y+LPE I KM+KL+VLIITNY GFH ++L+N E+LG L NL RIRL ++SVPS
Sbjct: 476 LNLHTEKYTLPECIKKMTKLKVLIITNYKGFHCAELDNFEILGCLPNLRRIRLHQVSVPS 655
Query: 586 FGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLK 645
L NL+KLSLY C T AF+ ++ ISD PNL+EL +DYCKDLV L +G+CDI SLK
Sbjct: 656 LCKLVNLRKLSLYFCETKQAFQSNTVSISDILPNLKELCVDYCKDLVTLPSGLCDITSLK 835
Query: 646 KLNVT 650
KL++T
Sbjct: 836 KLSIT 850
>CA920022 similar to PIR|T03031|T030 NBS-LRR type resistance protein - rice
(fragment), partial (8%)
Length = 706
Score = 268 bits (685), Expect = 6e-72
Identities = 137/190 (72%), Positives = 155/190 (81%)
Frame = -1
Query: 231 SESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYAQMEKNSSDII 290
SE LVEK +FQ++ +KILVT VAF RF T ILKPLA ED+VTLF HY ++EKNSS I
Sbjct: 703 SEDLVEKFKFQISDYKILVTQGVAFSRFDKTFILKPLAQEDSVTLFRHYTEVEKNSSKIP 524
Query: 291 DKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSILDSNTELLTRLQK 350
DK+L+EKVV C GLPL IKVIATS + RP++LW KIVKELS+G SILDSNTELL RLQK
Sbjct: 523 DKDLIEKVVEHCKGLPLAIKVIATSFRYRPYELWEKIVKELSRGRSILDSNTELLIRLQK 344
Query: 351 IFDVLEDNPKIMECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLGIMN 410
I DVLEDN ECFMDLALFPED RIPVAAL+DMWAELY LDD+G +AM+IINKL MN
Sbjct: 343 ILDVLEDNAISKECFMDLALFPEDQRIPVAALIDMWAELYGLDDDGKEAMDIINKLDSMN 164
Query: 411 LANVIIPRKN 420
LANV+I R N
Sbjct: 163 LANVLIARYN 134
>BF643326 weakly similar to GP|11994216|db disease resistance comples protein
{Arabidopsis thaliana}, partial (2%)
Length = 486
Score = 204 bits (519), Expect(2) = 3e-56
Identities = 102/125 (81%), Positives = 112/125 (89%)
Frame = +2
Query: 581 ISVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICD 640
ISVPSFGT+KNLKKLSLYMCNT LAFEKGSILISD FPNLE+L+IDY KD+V L G+CD
Sbjct: 2 ISVPSFGTMKNLKKLSLYMCNTRLAFEKGSILISDLFPNLEDLSIDYSKDMVALPNGVCD 181
Query: 641 IISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNC 700
I SLKKL++TNCHKLSSLPQDIGKL NLELLSL SCTDL +P SIG+LLNL+ LDISNC
Sbjct: 182 IASLKKLSITNCHKLSSLPQDIGKLMNLELLSLISCTDLVELPDSIGRLLNLRLLDISNC 361
Query: 701 ISLSS 705
ISLSS
Sbjct: 362 ISLSS 376
Score = 33.5 bits (75), Expect(2) = 3e-56
Identities = 13/18 (72%), Positives = 17/18 (94%)
Frame = +3
Query: 706 LPEEFGNLCNLKNLDMAT 723
LPE+FGNLCNL+NL M++
Sbjct: 378 LPEDFGNLCNLRNLYMSS 431
>BG646612 weakly similar to GP|9758140|dbj| disease resistance protein-like
{Arabidopsis thaliana}, partial (7%)
Length = 735
Score = 169 bits (428), Expect(2) = 3e-53
Identities = 103/155 (66%), Positives = 111/155 (71%), Gaps = 26/155 (16%)
Frame = +1
Query: 8 YYPYNKI**CLEQNI*NS*NSTKEPKFKAIV*VNLKGFISRCTRYKALQ*TLGSTQRRNK 67
YYPYNKI *CLEQNI NS*NSTK PK KAIV VNL+GF S +R+KA+Q*T GS+QRRN+
Sbjct: 22 YYPYNKIS*CLEQNISNS*NSTKAPKCKAIVEVNLEGFNSCGSRHKAIQ*TFGSSQRRNQ 201
Query: 68 *F-----DRRKCMQIFF*E*FLRC**EPISYCK*CRRDTLQGQRNS*ASQ---------- 112
RRKCMQ+FF*E*FL C**EPISYCK*C RDTL GQRNS*ASQ
Sbjct: 202 DSIRRK*CRRKCMQMFF*E*FLCC**EPISYCK*C*RDTL*GQRNS*ASQL*NF*TQIQ* 381
Query: 113 -----------PRKSKIHCWVGYTI*QVENGASEG 136
KSKIHCWVGYTI*QVENGA +G
Sbjct: 382 S*TTFQASFRCS*KSKIHCWVGYTI*QVENGAYKG 486
Score = 58.5 bits (140), Expect(2) = 3e-53
Identities = 37/56 (66%), Positives = 40/56 (71%), Gaps = 5/56 (8%)
Frame = +3
Query: 136 GVVAPHLC*LV*EDWGKPLWLQSFVGMKKLMEN----IIFVTFSK-TPMLKTIVER 186
G+VAP LC* *ED GKPLWLQSFVG+KKLM N IIFVT K T + IVER
Sbjct: 483 GMVAPPLC*RA*ED*GKPLWLQSFVGIKKLMVNSRKTIIFVTLLKNTHG*RPIVER 650
Score = 33.1 bits (74), Expect = 0.42
Identities = 22/39 (56%), Positives = 25/39 (63%), Gaps = 2/39 (5%)
Frame = +2
Query: 176 KTPMLKTIV-ERIHQHCGYSVP-EFQSDEDAVNKLGLLL 212
K P LKT E+ CGY VP F+ DE+AVNKL LLL
Sbjct: 617 KHPWLKTYCREK*LNICGYPVP*VFKVDENAVNKLELLL 733
>AW586679 similar to GP|9758140|dbj| disease resistance protein-like
{Arabidopsis thaliana}, partial (4%)
Length = 481
Score = 149 bits (375), Expect = 5e-36
Identities = 78/150 (52%), Positives = 102/150 (68%)
Frame = +3
Query: 152 KPLWLQSFVGMKKLMENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLL 211
K W Q F G K NI FVT S+TP LK IV+ + + CG VP+F ++EDA+N+LG L
Sbjct: 36 KLCWEQDFKG--KFGGNIFFVTVSETPNLKNIVKTLFEQCGRPVPDFTNEEDAINQLGHL 209
Query: 212 LKKVEGSPLLLVLDDVWPSSESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHED 271
L + E S +LLVLDDVWP SESLVEK F++ +KILVTSRV F RF T C L PL +
Sbjct: 210 LSQFERSQILLVLDDVWPGSESLVEKFTFKLPDYKILVTSRVGFRRFGTPCQLNPLDEDP 389
Query: 272 AVTLFHHYAQMEKNSSDIIDKNLVEKVVRS 301
A +LF HYAQ+ S + D +LV+++V++
Sbjct: 390 AASLFRHYAQLHHIISYMPDGDLVDEIVKA 479
>BG647778 weakly similar to PIR|T48468|T484 disease resistance-like protein -
Arabidopsis thaliana, partial (4%)
Length = 660
Score = 120 bits (302), Expect(3) = 2e-29
Identities = 71/150 (47%), Positives = 94/150 (62%), Gaps = 3/150 (2%)
Frame = +2
Query: 164 KLMENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLKKV-EGSPLLL 222
K ENI+F T SK P LK IV+ + +HCG+ P D+DAV L LL K+ E P++L
Sbjct: 173 KFGENILFFTVSKIPNLKNIVQTLFRHCGHDEPCLIDDDDAVKHLRSLLTKIGESCPMML 352
Query: 223 VLDDVWPSSESLVEKLQFQMTGFKILVTSRVAFPRFSTTCILKPLAHEDAVTLFHHYA-- 280
VLD+V P SESLVE Q Q+ KIL+TSRV FPRFS + LKPL +DAVTLF +A
Sbjct: 353 VLDNVCPGSESLVEDFQVQVPDCKILITSRVEFPRFS-SLFLKPLRDDDAVTLFGSFALP 529
Query: 281 QMEKNSSDIIDKNLVEKVVRSCNGLPLTIK 310
++ + + V++V + C G PL +K
Sbjct: 530 NDATRATYVPAEKYVKQVAKGCWGSPLALK 619
Score = 25.4 bits (54), Expect(3) = 2e-29
Identities = 16/33 (48%), Positives = 19/33 (57%)
Frame = +3
Query: 137 VVAPHLC*LV*EDWGKPLWLQSFVGMKKLMENI 169
VV +L * V D K LWL+ VGM KL N+
Sbjct: 81 VVCLYLF*PVWLDLAKLLWLRCSVGMTKLEVNL 179
Score = 22.3 bits (46), Expect(3) = 2e-29
Identities = 6/12 (50%), Positives = 9/12 (75%)
Frame = +1
Query: 119 HCWVGYTI*QVE 130
HCW+G++ VE
Sbjct: 28 HCWIGFSFDSVE 63
>TC92621 similar to GP|7107252|gb|AAF36340.1| unknown {Pennisetum glaucum},
partial (10%)
Length = 827
Score = 97.1 bits (240), Expect(3) = 2e-27
Identities = 46/67 (68%), Positives = 56/67 (82%)
Frame = +2
Query: 164 KLMENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLKKVEGSPLLLV 223
K ENIIFVTFSKTPMLK +V+R+ +H GY VPE+QSDE AVN +G LL+K+EGSP+L V
Sbjct: 608 KFKENIIFVTFSKTPMLKIMVQRLFEHGGYPVPEYQSDEXAVNGMGNLLRKIEGSPILXV 787
Query: 224 LDDVWPS 230
LDDV P+
Sbjct: 788 LDDVGPA 808
Score = 37.4 bits (85), Expect(3) = 2e-27
Identities = 19/33 (57%), Positives = 22/33 (66%)
Frame = +3
Query: 136 GVVAPHLC*LV*EDWGKPLWLQSFVGMKKLMEN 168
G+V C*LV* +W KPLWLQS V + KL N
Sbjct: 513 GMVDLLFC*LV*VEWEKPLWLQSCVWINKLRAN 611
Score = 27.3 bits (59), Expect(3) = 2e-27
Identities = 11/22 (50%), Positives = 15/22 (68%)
Frame = +1
Query: 115 KSKIHCWVGYTI*QVENGASEG 136
KS I+CW G+ I E+G S+G
Sbjct: 451 KS*IYCWFGFAID*AEDGTSQG 516
>BE319778
Length = 378
Score = 119 bits (298), Expect = 4e-27
Identities = 57/60 (95%), Positives = 58/60 (96%)
Frame = +2
Query: 418 RKNASDTDNNNYNNHFIILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQGL 477
RKNASDTDNNNYNNHFIILHDILRELGIYQS KEPFEQRKRLIID+N NKSGLAEKQQGL
Sbjct: 197 RKNASDTDNNNYNNHFIILHDILRELGIYQSAKEPFEQRKRLIIDINKNKSGLAEKQQGL 376
Score = 41.6 bits (96), Expect = 0.001
Identities = 20/20 (100%), Positives = 20/20 (100%)
Frame = +1
Query: 399 AMEIINKLGIMNLANVIIPR 418
AMEIINKLGIMNLANVIIPR
Sbjct: 1 AMEIINKLGIMNLANVIIPR 60
>CB893781 homologue to PIR|C64504|C645 hypothetical protein MJ1637 -
Methanococcus jannaschii, partial (2%)
Length = 473
Score = 67.8 bits (164), Expect(2) = 6e-23
Identities = 31/31 (100%), Positives = 31/31 (100%)
Frame = +3
Query: 136 GVVAPHLC*LV*EDWGKPLWLQSFVGMKKLM 166
GVVAPHLC*LV*EDWGKPLWLQSFVGMKKLM
Sbjct: 177 GVVAPHLC*LV*EDWGKPLWLQSFVGMKKLM 269
Score = 58.5 bits (140), Expect(2) = 6e-23
Identities = 36/60 (60%), Positives = 36/60 (60%), Gaps = 16/60 (26%)
Frame = +1
Query: 93 YCK*CRRDTLQGQRNS*ASQPR----------------KSKIHCWVGYTI*QVENGASEG 136
Y K* RD L QRNS*ASQP KSKIHCWVGYTI QVENGASEG
Sbjct: 1 YSK*S*RDAL*DQRNS*ASQP*IQ*RWTTFQASFQCS*KSKIHCWVGYTIYQVENGASEG 180
>TC79479 similar to GP|12056928|gb|AAG48132.1 putative resistance protein
{Glycine max}, partial (21%)
Length = 1087
Score = 87.8 bits (216), Expect = 1e-17
Identities = 66/201 (32%), Positives = 103/201 (50%)
Frame = +1
Query: 542 KMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISVPSFGTLKNLKKLSLYMCN 601
KM+ L+ LII + F N L SL+ LE S+PS+ K L L L +
Sbjct: 97 KMTGLQTLIIRSLCFAEGPKN---LPNSLRVLEWWGYPSQSLPSYFYPKKLAVLKLPHSS 267
Query: 602 TILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQD 661
F + S F N+ LN D CK ++ + +L++L++ +C L +
Sbjct: 268 ----FMSLELSKSKKFVNMTLLNFDECK-IITHIPDVSGAPNLERLSLDSCENLVEIHDS 432
Query: 662 IGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDM 721
+G L+ LE+L+L SC L +P L +L+HL++S+C SL S PE GN+ N+ +L +
Sbjct: 433 VGFLDKLEILNLGSCAKLRNLPPI--HLTSLQHLNLSHCSSLVSFPEILGNMKNITSLSL 606
Query: 722 ATCASIELPFSVVNLQNLKTI 742
A E P+S+ NL LK++
Sbjct: 607 EYTAIREFPYSIGNLPRLKSL 669
>BQ148232 similar to GP|9758141|dbj| disease resistance protein-like
{Arabidopsis thaliana}, partial (3%)
Length = 684
Score = 77.8 bits (190), Expect = 1e-14
Identities = 35/54 (64%), Positives = 45/54 (82%)
Frame = +3
Query: 195 VPEFQSDEDAVNKLGLLLKKVEGSPLLLVLDDVWPSSESLVEKLQFQMTGFKIL 248
+PEFQSDEDA+N+LGLL K+ P+LL+LDDVWP SESLV+K +FQ+ +KIL
Sbjct: 102 MPEFQSDEDAINELGLLFWKISIYPILLILDDVWPGSESLVDKFRFQLPDYKIL 263
>AW683902 weakly similar to PIR|T48468|T484 disease resistance-like protein -
Arabidopsis thaliana, partial (4%)
Length = 262
Score = 67.4 bits (163), Expect(2) = 4e-13
Identities = 29/48 (60%), Positives = 39/48 (80%)
Frame = +1
Query: 618 PNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKL 665
PNL EL+IDYC DL+ L +C+I +LKKL++TNCHKLS +P+DI K+
Sbjct: 1 PNLVELSIDYCNDLIKLPDELCNITTLKKLSITNCHKLSLMPRDIWKV 144
Score = 36.2 bits (82), Expect = 0.049
Identities = 17/43 (39%), Positives = 24/43 (55%)
Frame = +1
Query: 667 NLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEE 709
NL LS+ C DL +P + + LK L I+NC LS +P +
Sbjct: 4 NLVELSIDYCNDLIKLPDELCNITTLKKLSITNCHKLSLMPRD 132
Score = 25.8 bits (55), Expect(2) = 4e-13
Identities = 11/14 (78%), Positives = 12/14 (85%)
Frame = +2
Query: 663 GKLENLELLSLSSC 676
GKLENLE+L L SC
Sbjct: 137 GKLENLEVLRLCSC 178
>TC77571 homologue to GP|20804846|dbj|BAB92528. putative rust resistance
protein {Oryza sativa (japonica cultivar-group)},
partial (1%)
Length = 954
Score = 56.6 bits (135), Expect(2) = 1e-11
Identities = 32/66 (48%), Positives = 39/66 (58%)
Frame = +2
Query: 164 KLMENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLGLLLKKVEGSPLLLV 223
K ENIIFVTFSK P + + + + N LGLLL+K+EGSP+LLV
Sbjct: 752 KFKENIIFVTFSKHPC*RLL*RDYLNIADIRCLSTKVMKMRENGLGLLLRKIEGSPILLV 931
Query: 224 LDDVWP 229
LDDVWP
Sbjct: 932 LDDVWP 949
Score = 55.1 bits (131), Expect = 1e-07
Identities = 40/105 (38%), Positives = 50/105 (47%), Gaps = 10/105 (9%)
Frame = +1
Query: 115 KSKIHCWVGYTI*QVENGASEGVVAPHLC*LV*EDWGKPLWL-QSFVGMKK--------- 164
KS I+CW G+ I VE G S+ DW W ++ G K
Sbjct: 595 KS*IYCWFGFAID*VEGGNSQRR*IYSFV-----DWSG--WNGKNNSGYKAVFG*SS*G* 753
Query: 165 LMENIIFVTFSKTPMLKTIVERIHQHCGYSVPEFQSDEDAVNKLG 209
+ KTPMLK IVER+ +HCGY VPE+QSDEDA +G
Sbjct: 754 IQGKHYICNLLKTPMLKIIVERLFEHCGYPVPEYQSDEDAGKWIG 888
Score = 31.2 bits (69), Expect(2) = 1e-11
Identities = 16/26 (61%), Positives = 17/26 (64%)
Frame = +3
Query: 143 C*LV*EDWGKPLWLQSFVGMKKLMEN 168
C*LV +W K LWLQS V M KL N
Sbjct: 678 C*LVWVEWEKQLWLQSCVWMIKLRVN 755
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.333 0.145 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,076,712
Number of Sequences: 36976
Number of extensions: 358632
Number of successful extensions: 3500
Number of sequences better than 10.0: 302
Number of HSP's better than 10.0 without gapping: 1911
Number of HSP's successfully gapped in prelim test: 168
Number of HSP's that attempted gapping in prelim test: 1207
Number of HSP's gapped (non-prelim): 2323
length of query: 776
length of database: 9,014,727
effective HSP length: 104
effective length of query: 672
effective length of database: 5,169,223
effective search space: 3473717856
effective search space used: 3473717856
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148347.4


