

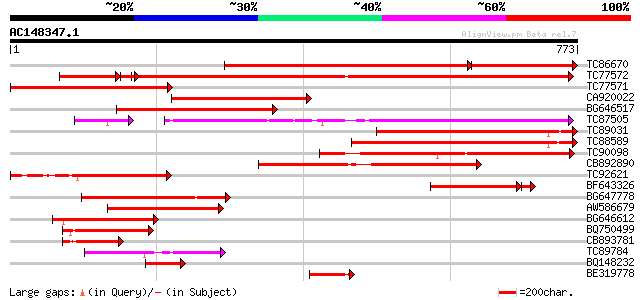
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148347.1 + phase: 0
(773 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86670 weakly similar to GP|9758141|dbj|BAB08633.1 disease resi... 667 0.0
TC77572 similar to GP|9558523|dbj|BAB03441.1 ESTs AU078742(C1188... 856 0.0
TC77571 homologue to GP|20804846|dbj|BAB92528. putative rust res... 380 e-105
CA920022 similar to PIR|T03031|T030 NBS-LRR type resistance prot... 368 e-102
BG646517 334 7e-92
TC87505 weakly similar to PIR|T05981|T05981 hypothetical protein... 314 9e-86
TC89031 weakly similar to GP|9758141|dbj|BAB08633.1 disease resi... 313 2e-85
TC88589 weakly similar to GP|9758140|dbj|BAB08632.1 disease resi... 313 2e-85
TC90098 weakly similar to GP|9758140|dbj|BAB08632.1 disease resi... 291 6e-79
CB892890 weakly similar to GP|9758141|dbj| disease resistance pr... 286 3e-77
TC92621 similar to GP|7107252|gb|AAF36340.1| unknown {Pennisetum... 256 2e-68
BF643326 weakly similar to GP|11994216|db disease resistance com... 228 1e-64
BG647778 weakly similar to PIR|T48468|T484 disease resistance-li... 192 3e-49
AW586679 similar to GP|9758140|dbj| disease resistance protein-l... 184 1e-46
BG646612 weakly similar to GP|9758140|dbj| disease resistance pr... 133 3e-31
BQ750499 similar to OMNI|NT01MC4764 transport protein HasD puta... 127 2e-29
CB893781 homologue to PIR|C64504|C645 hypothetical protein MJ163... 94 3e-19
TC89784 weakly similar to PIR|T05981|T05981 hypothetical protein... 89 5e-18
BQ148232 similar to GP|9758141|dbj| disease resistance protein-l... 84 2e-16
BE319778 82 6e-16
>TC86670 weakly similar to GP|9758141|dbj|BAB08633.1 disease resistance
protein-like {Arabidopsis thaliana}, partial (7%)
Length = 2417
Score = 667 bits (1722), Expect(2) = 0.0
Identities = 337/337 (100%), Positives = 337/337 (100%)
Frame = +3
Query: 294 KGLPLAIKVIATSFRYRPYELWEKIVKELSRGRSILDSNTELLIRLQKILDVLEDNAISK 353
KGLPLAIKVIATSFRYRPYELWEKIVKELSRGRSILDSNTELLIRLQKILDVLEDNAISK
Sbjct: 3 KGLPLAIKVIATSFRYRPYELWEKIVKELSRGRSILDSNTELLIRLQKILDVLEDNAISK 182
Query: 354 ECFMDLALFPEDQRIPVAALIDMWAELYGLDDDGKEAMDIINKLDSMNLANVLIARKNAS 413
ECFMDLALFPEDQRIPVAALIDMWAELYGLDDDGKEAMDIINKLDSMNLANVLIARKNAS
Sbjct: 183 ECFMDLALFPEDQRIPVAALIDMWAELYGLDDDGKEAMDIINKLDSMNLANVLIARKNAS 362
Query: 414 DTENYYYNNHFIVLHDLLRELGNYQNTQEPIEQRKRQLINTNESKCDQRLREKQQGTMAH 473
DTENYYYNNHFIVLHDLLRELGNYQNTQEPIEQRKRQLINTNESKCDQRLREKQQGTMAH
Sbjct: 363 DTENYYYNNHFIVLHDLLRELGNYQNTQEPIEQRKRQLINTNESKCDQRLREKQQGTMAH 542
Query: 474 ILSKLIGWFDKPKPQKVPARTLSISIDETCASDWSQVQPALAEVLILNLQTKQYTFPELM 533
ILSKLIGWFDKPKPQKVPARTLSISIDETCASDWSQVQPALAEVLILNLQTKQYTFPELM
Sbjct: 543 ILSKLIGWFDKPKPQKVPARTLSISIDETCASDWSQVQPALAEVLILNLQTKQYTFPELM 722
Query: 534 EKMNKLKALIVINHGLRPSELNNLELLSSLSNLKRIRLERISVPSFGTLKNLKKLSLYMC 593
EKMNKLKALIVINHGLRPSELNNLELLSSLSNLKRIRLERISVPSFGTLKNLKKLSLYMC
Sbjct: 723 EKMNKLKALIVINHGLRPSELNNLELLSSLSNLKRIRLERISVPSFGTLKNLKKLSLYMC 902
Query: 594 NTRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNG 630
NTRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNG
Sbjct: 903 NTRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNG 1013
Score = 290 bits (742), Expect(2) = 0.0
Identities = 144/144 (100%), Positives = 144/144 (100%)
Frame = +1
Query: 630 GVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLD 689
GVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLD
Sbjct: 1012 GVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLD 1191
Query: 690 ISNCISLSSLPEDFGNLCNLRNLDMTSCASCELPFSVVNLQNLKVTCDEETAASWESFQS 749
ISNCISLSSLPEDFGNLCNLRNLDMTSCASCELPFSVVNLQNLKVTCDEETAASWESFQS
Sbjct: 1192 ISNCISLSSLPEDFGNLCNLRNLDMTSCASCELPFSVVNLQNLKVTCDEETAASWESFQS 1371
Query: 750 MISNLTIEVPHVEVNLNWLHANRS 773
MISNLTIEVPHVEVNLNWLHANRS
Sbjct: 1372 MISNLTIEVPHVEVNLNWLHANRS 1443
>TC77572 similar to GP|9558523|dbj|BAB03441.1 ESTs AU078742(C11888)
C26225(C11888) correspond to a region of the predicted
gene.~Similar to Oryza, partial (5%)
Length = 2535
Score = 856 bits (2211), Expect(2) = 0.0
Identities = 431/603 (71%), Positives = 501/603 (82%), Gaps = 1/603 (0%)
Frame = +3
Query: 167 KTPMLKIIVERLFEHCGYPVPEYQSDEDAVNGLGLLLRKIEGSPILLVLDDVWPGSEDLV 226
K P ++ VER+ +HCGYPVPE+QSDEDAVN LGLLL+K+EGSP+LLVLDDVWP SE LV
Sbjct: 648 KHPC*RLFVERIHQHCGYPVPEFQSDEDAVNRLGLLLKKVEGSPLLLVLDDVWPISEPLV 827
Query: 227 EKFKFQISDYKILVTSRVAFSRFDKTFILKPLAQEDSVTLFRHYTEVEKNSSKIPDKDLI 286
EK +FQISD+KILVTSRVAF RF T ILKPLA E++VTLF HY ++EKNSS I +K+L+
Sbjct: 828 EKIQFQISDFKILVTSRVAFPRFSTTCILKPLAHEEAVTLFHHYAQMEKNSSDIINKNLV 1007
Query: 287 EKVVEHCKGLPLAIKVIATSFRYRPYELWEKIVKELSRGRSILDSNTELLIRLQKILDVL 346
EKVV C+GLPL IKVIATS + RP ++W KI KELS+G SILDSNT+LL RLQKI DVL
Sbjct: 1008 EKVVRSCQGLPLTIKVIATSLKNRPRDMWRKIGKELSQGHSILDSNTDLLTRLQKIFDVL 1187
Query: 347 EDNAISKECFMDLALFPEDQRIPVAALIDMWAELYGLDDDGKEAMDIINKLDSMNLANVL 406
EDN ECFMD+ALFPED RIPVAAL+DMWAELY LDD+G +AM+IINKL MNLANV+
Sbjct: 1188 EDNPTIMECFMDIALFPEDHRIPVAALVDMWAELYRLDDNGIQAMEIINKLGIMNLANVI 1367
Query: 407 IARKNASDTENYYYNNHFIVLHDLLRELGNYQNTQEPIEQRKRQLINTNESKCDQRLREK 466
I RKNASDT+N YNNHFI+LHD+LRELG YQ+ +EP EQRKR +I+ N++K L EK
Sbjct: 1368 IPRKNASDTDNNNYNNHFIILHDILRELGIYQSAKEPFEQRKRLIIDINKNK--SGLAEK 1541
Query: 467 QQGTMAHILSKLIGWFDKPKPQKVPARTLSISIDETCASDWSQVQPALAEVLILNLQTKQ 526
QQG M ILSK + K PQ++ AR LS+S DETCA DWSQ++PA EVLILNL TKQ
Sbjct: 1542 QQGLMTCILSKFMRLCVKRNPQQLTARILSVSADETCAFDWSQMEPAQVEVLILNLHTKQ 1721
Query: 527 YTFPELMEKMNKLKALIVINHGLRPSELNNLELLSSLSNLKRIRLERISVPSFGTLKNLK 586
Y+ PE + KM+KL+ LI+ N+ PS+LNN+ELL SL NL+RIRLERI VPSFGTLKNLK
Sbjct: 1722 YSLPEWIAKMSKLRVLIITNYDFHPSKLNNIELLGSLQNLERIRLERIYVPSFGTLKNLK 1901
Query: 587 KLSLYMCNTRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNGVCDIISLKKLSITNCH 646
KLSLYMCNT LAFEKGSILISD FPNLE+L++DYCKD+ LP G+CDI LKKL +TNCH
Sbjct: 1902 KLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLPIGICDIFLLKKLRVTNCH 2081
Query: 647 KLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNCISLSSLPEDFGNL 706
KLS LPQ+IGKLENLELLSL SCTDL +P SIG+L NL+ LDISNCISLSSLPE+FGNL
Sbjct: 2082 KLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNL 2261
Query: 707 CNLRNLDMTSCASCELPFSVVNLQNLK-VTCDEETAASWESFQSMISNLTIEVPHVEVNL 765
CNL+NLDM SCAS ELPFSVVNLQNLK +TCDEETAA+WE FQ M+ N+ IEV HV+VNL
Sbjct: 2262 CNLKNLDMASCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLPNMKIEVLHVDVNL 2441
Query: 766 NWL 768
NWL
Sbjct: 2442 NWL 2450
Score = 91.7 bits (226), Expect(2) = 0.0
Identities = 47/84 (55%), Positives = 60/84 (70%)
Frame = +1
Query: 68 DVEGKLRELLKILDKENFGKKISGSILKGPFDVPANPEFTVGLDLQLIKLKVEILREGRS 127
DVE L + +IL+ N G + K PF+VP N +FTVGLD+ KLK+E+LR G S
Sbjct: 349 DVEETLYKTREILELLNHEFNEDGQLFKRPFNVPENQKFTVGLDIPFSKLKMELLRGGSS 528
Query: 128 TLLLTGLGGMGKTTLATKLCLDDQ 151
TL+LTGLGG+GKTTLATKLC D++
Sbjct: 529 TLVLTGLGGLGKTTLATKLCWDER 600
Score = 45.8 bits (107), Expect = 6e-05
Identities = 21/27 (77%), Positives = 23/27 (84%)
Frame = +2
Query: 151 QVKGKFKENIIFVTFSKTPMLKIIVER 177
+V GKFKENIIFVTFSKTPMLK I +
Sbjct: 599 EVNGKFKENIIFVTFSKTPMLKTICRK 679
>TC77571 homologue to GP|20804846|dbj|BAB92528. putative rust resistance
protein {Oryza sativa (japonica cultivar-group)},
partial (1%)
Length = 954
Score = 380 bits (976), Expect = e-105
Identities = 193/221 (87%), Positives = 198/221 (89%)
Frame = +2
Query: 1 MGETTELDVISSPQFQQRFKHDQKNLCCIKWFSLVKRCIFVVFYKKKNKDDFDSCVGDDD 60
MGETTELDVISSPQFQQRFKHDQKNLCCIKWFSLVKRCIFVVFYKKKNKDDFDSCVGDDD
Sbjct: 290 MGETTELDVISSPQFQQRFKHDQKNLCCIKWFSLVKRCIFVVFYKKKNKDDFDSCVGDDD 469
Query: 61 DKQKMGKDVEGKLRELLKILDKENFGKKISGSILKGPFDVPANPEFTVGLDLQLIKLKVE 120
DKQKMGKDVEGKLRELLKILDKENFGKKISGSILKGPFDVPANPEFTVGLDLQLIKLKVE
Sbjct: 470 DKQKMGKDVEGKLRELLKILDKENFGKKISGSILKGPFDVPANPEFTVGLDLQLIKLKVE 649
Query: 121 ILREGRSTLLLTGLGGMGKTTLATKLCLDDQVKGKFKENIIFVTFSKTPMLKIIVERLFE 180
ILREGRSTLLLTGLGGMGKTTLATKLCLDDQVKGKFKENIIFVTFSK P +++
Sbjct: 650 ILREGRSTLLLTGLGGMGKTTLATKLCLDDQVKGKFKENIIFVTFSKHPC*RLL*RDYLN 829
Query: 181 HCGYPVPEYQSDEDAVNGLGLLLRKIEGSPILLVLDDVWPG 221
+ + NGLGLLLRKIEGSPILLVLDDVWPG
Sbjct: 830 IADIRCLSTKVMKMRENGLGLLLRKIEGSPILLVLDDVWPG 952
>CA920022 similar to PIR|T03031|T030 NBS-LRR type resistance protein - rice
(fragment), partial (8%)
Length = 706
Score = 368 bits (945), Expect = e-102
Identities = 188/191 (98%), Positives = 188/191 (98%)
Frame = -1
Query: 221 GSEDLVEKFKFQISDYKILVTSRVAFSRFDKTFILKPLAQEDSVTLFRHYTEVEKNSSKI 280
GSEDLVEKFKFQISDYKILVT VAFSRFDKTFILKPLAQEDSVTLFRHYTEVEKNSSKI
Sbjct: 706 GSEDLVEKFKFQISDYKILVTQGVAFSRFDKTFILKPLAQEDSVTLFRHYTEVEKNSSKI 527
Query: 281 PDKDLIEKVVEHCKGLPLAIKVIATSFRYRPYELWEKIVKELSRGRSILDSNTELLIRLQ 340
PDKDLIEKVVEHCKGLPLAIKVIATSFRYRPYELWEKIVKELSRGRSILDSNTELLIRLQ
Sbjct: 526 PDKDLIEKVVEHCKGLPLAIKVIATSFRYRPYELWEKIVKELSRGRSILDSNTELLIRLQ 347
Query: 341 KILDVLEDNAISKECFMDLALFPEDQRIPVAALIDMWAELYGLDDDGKEAMDIINKLDSM 400
KILDVLEDNAISKECFMDLALFPEDQRIPVAALIDMWAELYGLDDDGKEAMDIINKLDSM
Sbjct: 346 KILDVLEDNAISKECFMDLALFPEDQRIPVAALIDMWAELYGLDDDGKEAMDIINKLDSM 167
Query: 401 NLANVLIARKN 411
NLANVLIAR N
Sbjct: 166 NLANVLIARYN 134
>BG646517
Length = 668
Score = 334 bits (857), Expect = 7e-92
Identities = 167/220 (75%), Positives = 186/220 (83%)
Frame = +2
Query: 146 LCLDDQVKGKFKENIIFVTFSKTPMLKIIVERLFEHCGYPVPEYQSDEDAVNGLGLLLRK 205
LC D +V GKFKENIIFVTFSKTPMLK IVER+ EHCGYPVPE+QSDEDAVN L LLL+K
Sbjct: 2 LCWDQEVNGKFKENIIFVTFSKTPMLKTIVERIIEHCGYPVPEFQSDEDAVNKLELLLKK 181
Query: 206 IEGSPILLVLDDVWPGSEDLVEKFKFQISDYKILVTSRVAFSRFDKTFILKPLAQEDSVT 265
+EGSP+LLVLDDVWP SE LV+K +F+ISD+KILVTSRV+F RF T ILKPLA ED+VT
Sbjct: 182 VEGSPLLLVLDDVWPTSESLVKKLQFEISDFKILVTSRVSFPRFRTTCILKPLAHEDAVT 361
Query: 266 LFRHYTEVEKNSSKIPDKDLIEKVVEHCKGLPLAIKVIATSFRYRPYELWEKIVKELSRG 325
LF HY ++EKNSS I DK+L+EKVV CKGLPL IKVIATS R RP +LW KIV ELS+G
Sbjct: 362 LFHHYAQMEKNSSDIIDKNLVEKVVRSCKGLPLTIKVIATSLRNRPDDLWRKIVMELSQG 541
Query: 326 RSILDSNTELLIRLQKILDVLEDNAISKECFMDLALFPED 365
SILDSNTELL RLQKI DVLEDN ECFMD+ALFPED
Sbjct: 542 HSILDSNTELLTRLQKIFDVLEDNPTIMECFMDIALFPED 661
>TC87505 weakly similar to PIR|T05981|T05981 hypothetical protein F17M5.60 -
Arabidopsis thaliana, partial (45%)
Length = 2140
Score = 314 bits (804), Expect = 9e-86
Identities = 188/566 (33%), Positives = 318/566 (55%), Gaps = 8/566 (1%)
Frame = +3
Query: 211 ILLVLDDVWPGSEDLVEKFKFQISDYKILVTSRVAFSRFDKTFILKPLAQEDSVTLFRHY 270
IL++LDDVW S ++E+ F++ + K +V SR F F+ T+ ++ L ++D+++LF H+
Sbjct: 396 ILVILDDVW--SPSVLEQLVFRMPNCKFIVVSRFIFPIFNATYKVELLDKDDALSLFCHH 569
Query: 271 TEVEKNSSKIPDKDLIEKVVEHCKGLPLAIKVIATSFRYRPYELWEKIVKELSRGRSILD 330
+K+ +++L+++VV C LPLA+KVI S R + W + LS+G SI +
Sbjct: 570 AFGQKSIPFAANQNLVKQVVAECGNLPLALKVIGASLRDQNEMFWLSVKTRLSQGLSIDE 749
Query: 331 SNTELLIRLQKILDVLEDNAISKECFMDLALFPEDQRIPVAALIDMWAELYGLDDDGKEA 390
S LI I I KECF+DL FPED++IP+ LI+MW E++ + + EA
Sbjct: 750 SYERNLIDRMAISTNYLPEKI-KECFLDLCSFPEDKKIPLEVLINMWVEIHDIHET--EA 920
Query: 391 MDIINKLDSMNLANVLIARKNASDTENYYYNNHF---IVLHDLLRELGNYQNTQEPIEQR 447
I+ +L + NL ++ + Y++ F + HD+LR+L + + I QR
Sbjct: 921 YAIVVELSNKNLLTLVEEARAGG-----MYSSCFEISVTQHDILRDLALNLSNRGNINQR 1085
Query: 448 KRQLINTNESKCDQRLREKQQGTMAHILSKLIGWFDKPKPQKVPARTLSISIDETCASDW 507
+R ++ E + + + + D+P A+ +SI E SDW
Sbjct: 1086RRLVMPKREDN-------------GQLPKEWLRYADQP----FEAQIVSIHTGEMRKSDW 1214
Query: 508 SQVQPALAEVLILNLQTKQYTFPELMEKMNKLKALIVINHGLRPSELNNLELLSSLSNLK 567
++ AEVLI+N + +Y P + +M KL+AL+VINH + L+N+ + +L+NL+
Sbjct: 1215CNLEFPKAEVLIINFTSSEYFLPPFINRMPKLRALMVINHSTSYACLHNISVFKNLTNLR 1394
Query: 568 RIRLERISVPSFG--TLKNLKKLSLYMCNTRLAFEKGSILISDLFPNLEDLSMDYCKDMT 625
+ E++S+P +++L+KL + +C + E I+D+FPN+ +L++D+C+D+T
Sbjct: 1395SLWFEKVSIPHLSGIVMESLRKLFIVLCKINNSLEGKDSNIADIFPNISELTLDHCEDVT 1574
Query: 626 ALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNL 685
LP+ +C I SL+ LS+TNCH L+ LP E+G L LE+L L +C +L LP SI ++ L
Sbjct: 1575ELPSSICRIQSLQNLSLTNCHSLTRLPIELGSLRYLEILRLYACPNLRTLPPSICGMTRL 1754
Query: 686 RLLDISNCISLSSLPEDFGNLCNLRNLDMTSCAS-CELPFSVVNLQNLKV--TCDEETAA 742
+ +DI+ C+ L+S P+ G L NL +DM C +P S ++L CD+E +
Sbjct: 1755KYIDIAQCVYLASFPDAIGKLVNLEKIDMRECPMITNIPKSALSLNFFCSL*ICDDEVSW 1934
Query: 743 SWESFQSMISNLTIEVPHVEVNLNWL 768
W+ Q + N+ I+V +E +L+WL
Sbjct: 1935MWKEVQKVKLNVDIQVVEIEYDLDWL 2012
Score = 56.6 bits (135), Expect = 3e-08
Identities = 31/84 (36%), Positives = 48/84 (56%), Gaps = 3/84 (3%)
Frame = +2
Query: 89 ISGSILKGPFDVPANPEFTVGLDLQLIKLKVEILREGRSTLLL---TGLGGMGKTTLATK 145
+ G++ D + + L+ K KV + GR L +G+GG GKTTLA +
Sbjct: 23 VEGAVRSSEEDEGSLGNLDLSFGLEFGKNKVMEMVVGRKDFCLVGISGIGGSGKTTLARE 202
Query: 146 LCLDDQVKGKFKENIIFVTFSKTP 169
+C D+QV+G FKE I+F+T S++P
Sbjct: 203 ICRDEQVRGYFKERILFLTVSQSP 274
>TC89031 weakly similar to GP|9758141|dbj|BAB08633.1 disease resistance
protein-like {Arabidopsis thaliana}, partial (5%)
Length = 1294
Score = 313 bits (801), Expect = 2e-85
Identities = 162/279 (58%), Positives = 203/279 (72%), Gaps = 5/279 (1%)
Frame = +3
Query: 500 DETCASDWSQVQPALAEVLILNLQTKQYTFPELMEKMNKLKALIVINHGLRPSELNNLEL 559
DE+ +SDW +QP EVL+LNLQ+ QY+ P+ +KM+KLK LIV N+G SEL EL
Sbjct: 123 DESFSSDWCDMQPDEVEVLVLNLQSDQYSLPDFTDKMSKLKVLIVTNYGFHRSELIKFEL 302
Query: 560 LSSLSNLKRIRLERISVPSFGTLKNLKKLSLYMCNTRLAFEKGSILISDLFPNLEDLSMD 619
L LSNLKRIRLE++SVP LKNL+KLSL+MCNTR AFE SI ISD PNL +LS+D
Sbjct: 303 LGFLSNLKRIRLEKVSVPCLSILKNLQKLSLHMCNTRDAFENYSIQISDAMPNLVELSID 482
Query: 620 YCKDMTALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSI 679
YC D+ LP+G +I +LKK+SITNCHKLS +PQ+I KLENLE+L L SC+DLVE+ +S+
Sbjct: 483 YCNDLIKLPDGFSNITTLKKISITNCHKLSAIPQDIEKLENLEVLRLCSCSDLVEISESV 662
Query: 680 GRLSNLRLLDISNCISLSSLPEDFGNLCNLRNLDMTSCAS-CELPFSVVNLQNLK----V 734
L+ LR DIS+C+SLS LP D G+L L M C++ ELP+SV+NL N+K V
Sbjct: 663 SGLNKLRCFDISDCVSLSKLPNDIGDLKKLEKFYMKGCSNLSELPYSVINLGNVKHEIHV 842
Query: 735 TCDEETAASWESFQSMISNLTIEVPHVEVNLNWLHANRS 773
CDEE AA WE F + I NL I++P VE+NLNWLH RS
Sbjct: 843 ICDEEGAALWEHFPN-IPNLKIDMPKVEINLNWLHGTRS 956
>TC88589 weakly similar to GP|9758140|dbj|BAB08632.1 disease resistance
protein-like {Arabidopsis thaliana}, partial (12%)
Length = 1137
Score = 313 bits (801), Expect = 2e-85
Identities = 172/312 (55%), Positives = 211/312 (67%), Gaps = 5/312 (1%)
Frame = +1
Query: 467 QQGTMAHILSKLIGWFDKPKPQKVPARTLSISIDETCASDWSQVQPALAEVLILNLQTKQ 526
QQG +++ S + G K K KV AR L IS DE +SDW +QP AEVL+LNL++ Q
Sbjct: 25 QQGIISNWYSFITGMLVKQKQLKVAARILCISTDEIFSSDWCDMQPDKAEVLVLNLRSDQ 204
Query: 527 YTFPELMEKMNKLKALIVINHGLRPSELNNLELLSSLSNLKRIRLERISVPSFGTLKNLK 586
Y+ P+ +KM KLK LIV N+G SEL ELL SLSNLKRIRLE++SVP LKNL+
Sbjct: 205 YSLPDFTKKMRKLKVLIVTNYGFSRSELTKFELLGSLSNLKRIRLEKVSVPCLCILKNLR 384
Query: 587 KLSLYMCNTRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNGVCDIISLKKLSITNCH 646
KLSL+MC+T AFE SI ISD PNL +LS+DYC D+ LP C I +LKKLSITNCH
Sbjct: 385 KLSLHMCSTNNAFESCSIQISDAMPNLVELSIDYCNDLIKLPGEFCKITTLKKLSITNCH 564
Query: 647 KLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNCISLSSLPEDFGNL 706
K S +PQ+IGKL NLE+L L SC+DL E+P+S+ L+ LR LDIS+C++L LP + GNL
Sbjct: 565 KFSAMPQDIGKLVNLEVLRLCSCSDLKEIPESVADLNKLRCLDISDCVTLHILPNNIGNL 744
Query: 707 CNLRNLDMTSCAS-CELPFSVVNLQNLK----VTCDEETAASWESFQSMISNLTIEVPHV 761
L L M C++ ELP SV+N NLK V CDEE +A WE S I L I +P V
Sbjct: 745 QKLEKLYMKGCSNLSELPDSVINFGNLKHEMQVICDEEGSALWEHL-SNIPKLKIYMPKV 921
Query: 762 EVNLNWLHANRS 773
E NL WLH RS
Sbjct: 922 EHNLIWLHGIRS 957
>TC90098 weakly similar to GP|9758140|dbj|BAB08632.1 disease resistance
protein-like {Arabidopsis thaliana}, partial (16%)
Length = 1128
Score = 291 bits (745), Expect = 6e-79
Identities = 165/354 (46%), Positives = 222/354 (62%), Gaps = 7/354 (1%)
Frame = +1
Query: 423 HFIVLHDLLRELGNYQNTQEPIEQRKRQLINTNESKCDQRLREKQQGTMAHILSKLIGWF 482
H++ H LLR+L N+QE ++R R +I+T+ + L W+
Sbjct: 1 HYVTQHGLLRDLAIRDNSQEAEDKRNRLIIDTSANN-------------------LPSWW 123
Query: 483 DKPKPQKVPARTLSISIDETCASDWSQVQPALAEVLILNLQTKQYTFPELMEKMNKLKAL 542
+ AR LSIS DE S W +QP EVL+LNL+ K++ P M+KM KLKAL
Sbjct: 124 TSENEYHIKARILSISTDEIFTSKWCNLQPTEVEVLVLNLREKKFALPMFMKKMKKLKAL 303
Query: 543 IVINHGLRPSELNNLELLSSLSNLKRIRLERISVPSFGT----LKNLKKLSLYMCNTRLA 598
I+ N+ P+EL N E+L LSNLKRIRLE++SVP G LKNL+K S +MCN A
Sbjct: 304 IITNYDFFPAELENFEVLDHLSNLKRIRLEKVSVPFLGKTVLQLKNLQKCSFFMCNVNKA 483
Query: 599 FEKGSILISDLFPNLEDLSMDYCKDMTALPNGVCDIISLKKLSITNCHKLSLLPQEIGKL 658
FE +I S++ PNL +++ DYC DM LPN + +I+SLKKLSITNCHKL L + IGKL
Sbjct: 484 FENCTIEDSEMLPNLMEMNFDYC-DMVELPNVISNIVSLKKLSITNCHKLCALHEGIGKL 660
Query: 659 ENLELLSLISCTDLVELPDSIGRLSNLRLLDISNCISLSSLPEDFGNLCNLRNLDMTSCA 718
NLE L L SC+ L+ELP+SI L L+ L+IS CISLS LPE+ G L L L+M C+
Sbjct: 661 VNLESLRLSSCSGLLELPNSIRNLHVLKFLNISGCISLSQLPENIGELEKLEKLNMRGCS 840
Query: 719 S-CELPFSVVNLQNLK-VTC-DEETAASWESFQSMISNLTIEVPHVEVNLNWLH 769
+ ELP SV+ L+ LK V C DEETA WE +++++ +L +EV + NLN+L+
Sbjct: 841 NISELPSSVMELEGLKHVVCGDEETAEKWEPYKNILGDLKVEVVQEDFNLNFLY 1002
>CB892890 weakly similar to GP|9758141|dbj| disease resistance protein-like
{Arabidopsis thaliana}, partial (4%)
Length = 852
Score = 286 bits (731), Expect = 3e-77
Identities = 154/307 (50%), Positives = 208/307 (67%), Gaps = 3/307 (0%)
Frame = +2
Query: 340 QKILD-VLEDNAISKECFMDLALFPEDQRIPVAALIDMWAELYGLDDDGKEAMDIINKLD 398
QK+L+ L+DN I KECFMDL LF ED++IPVAALIDMW ELY LDDD + M+I+ KL
Sbjct: 2 QKVLENALQDNHIVKECFMDLGLFFEDKKIPVAALIDMWTELYDLDDDNIKGMNIVRKLA 181
Query: 399 SMNLANVLIARKNASDTENYYYNNHFIVLHDLLRELGNYQNTQEPIEQRKRQLINTNESK 458
+ +L ++++R+ + + +YYN+HF+ HDLL+E+ +Q QEP E RKR + + NE+
Sbjct: 182 NWHLVKLVVSREVTTHVD-HYYNHHFLTQHDLLKEISIHQARQEPFELRKRLIFDVNENS 358
Query: 459 CDQRLREKQQGTMAHILSKLIGWFDKPKPQKVPARTLSISIDETCASDWSQVQPAL-AEV 517
DQ + QQ T+A RTLSIS D+ SDWS V+ EV
Sbjct: 359 WDQ---QNQQNTIA--------------------RTLSISPDKILTSDWSNVEKIKQVEV 469
Query: 518 LILNLQTKQYTFPELMEKMNKLKALIVINH-GLRPSELNNLELLSSLSNLKRIRLERISV 576
LILNL T++YT PE ++KM KLK LI+ N+ G +EL+N E+L L NL+RIRL ++SV
Sbjct: 470 LILNLHTEKYTLPECIKKMTKLKVLIITNYKGFHCAELDNFEILGCLPNLRRIRLHQVSV 649
Query: 577 PSFGTLKNLKKLSLYMCNTRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNGVCDIIS 636
PS L NL+KLSLY C T+ AF+ ++ ISD+ PNL++L +DYCKD+ LP+G+CDI S
Sbjct: 650 PSLCKLVNLRKLSLYFCETKQAFQSNTVSISDILPNLKELCVDYCKDLVTLPSGLCDITS 829
Query: 637 LKKLSIT 643
LKKLSIT
Sbjct: 830 LKKLSIT 850
>TC92621 similar to GP|7107252|gb|AAF36340.1| unknown {Pennisetum glaucum},
partial (10%)
Length = 827
Score = 256 bits (654), Expect = 2e-68
Identities = 147/223 (65%), Positives = 159/223 (70%), Gaps = 3/223 (1%)
Frame = +2
Query: 1 MGETTELDVISSPQFQQRFKHDQKNLCCIKWFSLVKRCIFVVFYKKKNKDDFDSCVGDDD 60
MG ELD ISSPQ QQ KNLCC K+F C+ +KK +F S
Sbjct: 194 MGHNIELDAISSPQLQQ------KNLCCSKFFCWFHLCVSGFLHKK----NFGS------ 325
Query: 61 DKQKMGKDVEGKLRELLKILDKENFGKKISG---SILKGPFDVPANPEFTVGLDLQLIKL 117
D + D E +IL F KKISG SI KGPF VP NPEFTVGLD QLIK
Sbjct: 326 DSHAVAHDKPKTFYEFKEILF---FDKKISGYGVSIFKGPFGVPENPEFTVGLDSQLIKP 496
Query: 118 KVEILREGRSTLLLTGLGGMGKTTLATKLCLDDQVKGKFKENIIFVTFSKTPMLKIIVER 177
K+E+LR+GRSTLLLTGLGGMGKTTLATKLCLD QVKGKFKENIIFVTFSKTPMLKI+V+R
Sbjct: 497 KMELLRDGRSTLLLTGLGGMGKTTLATKLCLDQQVKGKFKENIIFVTFSKTPMLKIMVQR 676
Query: 178 LFEHCGYPVPEYQSDEDAVNGLGLLLRKIEGSPILLVLDDVWP 220
LFEH GYPVPEYQSDE AVNG+G LLRKIEGSPIL VLDDV P
Sbjct: 677 LFEHGGYPVPEYQSDEXAVNGMGNLLRKIEGSPILXVLDDVGP 805
>BF643326 weakly similar to GP|11994216|db disease resistance comples protein
{Arabidopsis thaliana}, partial (2%)
Length = 486
Score = 228 bits (582), Expect(2) = 1e-64
Identities = 116/125 (92%), Positives = 119/125 (94%)
Frame = +2
Query: 574 ISVPSFGTLKNLKKLSLYMCNTRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNGVCD 633
ISVPSFGT+KNLKKLSLYMCNTRLAFEKGSILISDLFPNLEDLS+DY KDM ALPNGVCD
Sbjct: 2 ISVPSFGTMKNLKKLSLYMCNTRLAFEKGSILISDLFPNLEDLSIDYSKDMVALPNGVCD 181
Query: 634 IISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNC 693
I SLKKLSITNCHKLS LPQ+IGKL NLELLSLISCTDLVELPDSIGRL NLRLLDISNC
Sbjct: 182 IASLKKLSITNCHKLSSLPQDIGKLMNLELLSLISCTDLVELPDSIGRLLNLRLLDISNC 361
Query: 694 ISLSS 698
ISLSS
Sbjct: 362 ISLSS 376
Score = 37.4 bits (85), Expect(2) = 1e-64
Identities = 16/18 (88%), Positives = 17/18 (93%)
Frame = +3
Query: 699 LPEDFGNLCNLRNLDMTS 716
LPEDFGNLCNLRNL M+S
Sbjct: 378 LPEDFGNLCNLRNLYMSS 431
>BG647778 weakly similar to PIR|T48468|T484 disease resistance-like protein -
Arabidopsis thaliana, partial (4%)
Length = 660
Score = 192 bits (489), Expect = 3e-49
Identities = 108/206 (52%), Positives = 136/206 (65%), Gaps = 3/206 (1%)
Frame = +2
Query: 99 DVPANPEFTVGLDLQLIKLKVEILREGRSTLLLTGLGGMGKTTLATKLCLDDQVKGKFKE 158
D P P+ TVGLD LI+LK +L G S L+LTGL G GKTTLAT LC DD+V+GKF E
Sbjct: 5 DAPVKPDLTVGLDFPLIQLKSWLLGSGVSVLVLTGLAGSGKTTLATLLCWDDKVRGKFGE 184
Query: 159 NIIFVTFSKTPMLKIIVERLFEHCGYPVPEYQSDEDAVNGLGLLLRKI-EGSPILLVLDD 217
NI+F T SK P LK IV+ LF HCG+ P D+DAV L LL KI E P++LVLD+
Sbjct: 185 NILFFTVSKIPNLKNIVQTLFRHCGHDEPCLIDDDDAVKHLRSLLTKIGESCPMMLVLDN 364
Query: 218 VWPGSEDLVEKFKFQISDYKILVTSRVAFSRFDKTFILKPLAQEDSVTLFRHYT--EVEK 275
V PGSE LVE F+ Q+ D KIL+TSRV F RF F LKPL +D+VTLF +
Sbjct: 365 VCPGSESLVEDFQVQVPDCKILITSRVEFPRFSSLF-LKPLRDDDAVTLFGSFALPNDAT 541
Query: 276 NSSKIPDKDLIEKVVEHCKGLPLAIK 301
++ +P + +++V + C G PLA+K
Sbjct: 542 RATYVPAEKYVKQVAKGCWGSPLALK 619
>AW586679 similar to GP|9758140|dbj| disease resistance protein-like
{Arabidopsis thaliana}, partial (4%)
Length = 481
Score = 184 bits (466), Expect = 1e-46
Identities = 92/158 (58%), Positives = 115/158 (72%)
Frame = +3
Query: 134 LGGMGKTTLATKLCLDDQVKGKFKENIIFVTFSKTPMLKIIVERLFEHCGYPVPEYQSDE 193
LGG GKTTLA KLC + KGKF NI FVT S+TP LK IV+ LFE CG PVP++ ++E
Sbjct: 3 LGGSGKTTLAKKLCWEQDFKGKFGGNIFFVTVSETPNLKNIVKTLFEQCGRPVPDFTNEE 182
Query: 194 DAVNGLGLLLRKIEGSPILLVLDDVWPGSEDLVEKFKFQISDYKILVTSRVAFSRFDKTF 253
DA+N LG LL + E S ILLVLDDVWPGSE LVEKF F++ DYKILVTSRV F RF
Sbjct: 183 DAINQLGHLLSQFERSQILLVLDDVWPGSESLVEKFTFKLPDYKILVTSRVGFRRFGTPC 362
Query: 254 ILKPLAQEDSVTLFRHYTEVEKNSSKIPDKDLIEKVVE 291
L PL ++ + +LFRHY ++ S +PD DL++++V+
Sbjct: 363 QLNPLDEDPAASLFRHYAQLHHIISYMPDGDLVDEIVK 476
>BG646612 weakly similar to GP|9758140|dbj| disease resistance protein-like
{Arabidopsis thaliana}, partial (7%)
Length = 735
Score = 133 bits (334), Expect = 3e-31
Identities = 80/153 (52%), Positives = 103/153 (67%), Gaps = 8/153 (5%)
Frame = +2
Query: 59 DDDKQKMGKDVEG---KLRELLKILDKENFGKKISGSI--LKGPFDVPANPEFTVGLDLQ 113
D+++ + DVE K RE+L++L+ E F K + + K PFDVP NP+FTVGLD+
Sbjct: 275 DENQSLIVNDVEETLYKAREILELLNYETFEHKFNEAKPPFKRPFDVPENPKFTVGLDIP 454
Query: 114 LIKLKVEILREGRSTLLLTGLGGMGKTTLATKLCLDDQVKGKFKEN-IIFVTFSKTPMLK 172
KLK+E++R+G STL+LTGLGG+GKTTLATKLC D +V GKFKEN I K P LK
Sbjct: 455 FSKLKMELIRDGSSTLVLTGLGGLGKTTLATKLCWDQEVNGKFKENHYICHPSQKHPWLK 634
Query: 173 IIV-ERLFEHCGYPVP-EYQSDEDAVNGLGLLL 203
E+ CGYPVP ++ DE+AVN L LLL
Sbjct: 635 TYCREK*LNICGYPVP*VFKVDENAVNKLELLL 733
>BQ750499 similar to OMNI|NT01MC4764 transport protein HasD putative
{Magnetococcus sp. MC-1}, partial (3%)
Length = 800
Score = 127 bits (319), Expect = 2e-29
Identities = 69/129 (53%), Positives = 91/129 (70%), Gaps = 4/129 (3%)
Frame = +1
Query: 72 KLRELLKILD----KENFGKKISGSILKGPFDVPANPEFTVGLDLQLIKLKVEILREGRS 127
K+ E+ KI + KEN G+ GS ++G P P+ +G+D L KLK+++L++G S
Sbjct: 409 KVDEISKIFEILMRKENLGQ-FDGSQIRGLCGAPEEPQ-CMGVDEPLNKLKIQLLKDGVS 582
Query: 128 TLLLTGLGGMGKTTLATKLCLDDQVKGKFKENIIFVTFSKTPMLKIIVERLFEHCGYPVP 187
L+LTGLGG GK+TLA KLC + Q+KGKF NI FVT SKTP LK IV+ LFEHCG VP
Sbjct: 583 VLVLTGLGGSGKSTLAKKLCWNPQIKGKFGGNIFFVTVSKTPNLKNIVQTLFEHCGLRVP 762
Query: 188 EYQSDEDAV 196
E+Q+DEDA+
Sbjct: 763 EFQTDEDAI 789
>CB893781 homologue to PIR|C64504|C645 hypothetical protein MJ1637 -
Methanococcus jannaschii, partial (2%)
Length = 473
Score = 93.6 bits (231), Expect = 3e-19
Identities = 48/84 (57%), Positives = 64/84 (76%)
Frame = +2
Query: 72 KLRELLKILDKENFGKKISGSILKGPFDVPANPEFTVGLDLQLIKLKVEILREGRSTLLL 131
K +E+L++L+ E F + G + K PF+VP N +FTVGLD+ KLK+E+LR G STL+L
Sbjct: 32 KTKEILELLNHE-FNE--DGQLFKRPFNVPENQKFTVGLDIPFTKLKMELLRGGSSTLVL 202
Query: 132 TGLGGMGKTTLATKLCLDDQVKGK 155
TGL G+GKTTLATKLC D++V GK
Sbjct: 203 TGLRGLGKTTLATKLCWDEEVNGK 274
>TC89784 weakly similar to PIR|T05981|T05981 hypothetical protein F17M5.60 -
Arabidopsis thaliana, partial (20%)
Length = 1100
Score = 89.4 bits (220), Expect = 5e-18
Identities = 61/202 (30%), Positives = 112/202 (55%), Gaps = 10/202 (4%)
Frame = +1
Query: 103 NPEFTVGLDLQLIKLKVEIL-REGRSTLLLTGLGGMGKTTLATKLCLDDQVKGKFKENII 161
N +VGLDL K+K ++ RE + + G+GG GKTTL ++C D+QV+ F E I+
Sbjct: 526 NLSLSVGLDLGKKKVKEMVMGREDLWVVGIHGIGGSGKTTLVKEICKDEQVRCYFNEKIL 705
Query: 162 FVTFSKTPMLKIIVERLFEHC--------GYPVPEYQSDEDAVNGLGLLLRKIEGSPILL 213
F+T S++P ++ + +++ H Y VP + + + + L+
Sbjct: 706 FLTVSQSPNVEQLRSKIWGHIMGNRNLNPNYVVPRWIPQFECRS----------EARTLV 855
Query: 214 VLDDVWPGSEDLVEKFKFQISDYKILVTSRVAF-SRFDKTFILKPLAQEDSVTLFRHYTE 272
VLDDVW S+ ++E+ +I K +V SR F + F T+ ++ L++ED+++LF H+
Sbjct: 856 VLDDVW--SQAVLEQLVCRIPGCKFVVVSRFQFPTIFSATYKVELLSEEDALSLFCHHAF 1029
Query: 273 VEKNSSKIPDKDLIEKVVEHCK 294
+K+ +++L+++VV C+
Sbjct: 1030GQKSIPLTANENLVKQVVSECE 1095
>BQ148232 similar to GP|9758141|dbj| disease resistance protein-like
{Arabidopsis thaliana}, partial (3%)
Length = 684
Score = 84.3 bits (207), Expect = 2e-16
Identities = 39/54 (72%), Positives = 46/54 (84%)
Frame = +3
Query: 186 VPEYQSDEDAVNGLGLLLRKIEGSPILLVLDDVWPGSEDLVEKFKFQISDYKIL 239
+PE+QSDEDA+N LGLL KI PILL+LDDVWPGSE LV+KF+FQ+ DYKIL
Sbjct: 102 MPEFQSDEDAINELGLLFWKISIYPILLILDDVWPGSESLVDKFRFQLPDYKIL 263
>BE319778
Length = 378
Score = 82.4 bits (202), Expect = 6e-16
Identities = 40/61 (65%), Positives = 49/61 (79%)
Frame = +2
Query: 409 RKNASDTENYYYNNHFIVLHDLLRELGNYQNTQEPIEQRKRQLINTNESKCDQRLREKQQ 468
RKNASDT+N YNNHFI+LHD+LRELG YQ+ +EP EQRKR +I+ N++K L EKQQ
Sbjct: 197 RKNASDTDNNNYNNHFIILHDILRELGIYQSAKEPFEQRKRLIIDINKNK--SGLAEKQQ 370
Query: 469 G 469
G
Sbjct: 371 G 373
Score = 32.3 bits (72), Expect = 0.71
Identities = 18/31 (58%), Positives = 21/31 (67%)
Frame = +1
Query: 390 AMDIINKLDSMNLANVLIARKNASDTENYYY 420
AM+IINKL MNLANV+I R +YYY
Sbjct: 1 AMEIINKLGIMNLANVIIPR-----YYDYYY 78
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.138 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,945,961
Number of Sequences: 36976
Number of extensions: 343207
Number of successful extensions: 2403
Number of sequences better than 10.0: 319
Number of HSP's better than 10.0 without gapping: 2047
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2243
length of query: 773
length of database: 9,014,727
effective HSP length: 104
effective length of query: 669
effective length of database: 5,169,223
effective search space: 3458210187
effective search space used: 3458210187
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148347.1


