

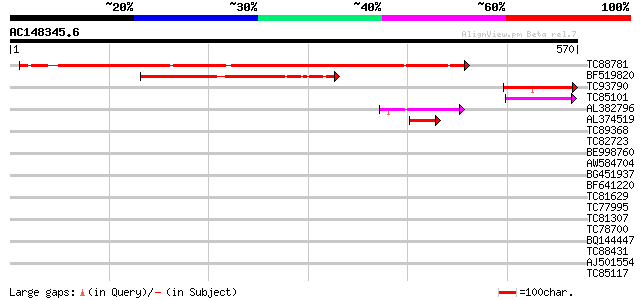
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148345.6 + phase: 0
(570 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88781 similar to GP|9369366|gb|AAF87115.1| F10A5.18 {Arabidops... 528 e-150
BF519820 weakly similar to GP|15809876|gb AT3g53950/F5K20_250 {A... 190 1e-48
TC93790 weakly similar to GP|9369366|gb|AAF87115.1| F10A5.18 {Ar... 77 2e-14
TC85101 similar to GP|15809876|gb|AAL06866.1 AT3g53950/F5K20_250... 54 2e-07
AL382796 weakly similar to GP|20161084|dbj putative glyoxal oxid... 53 3e-07
AL374519 similar to GP|15809876|gb| AT3g53950/F5K20_250 {Arabido... 47 3e-05
TC89368 similar to SP|O22160|TL15_ARATH Thylakoid lumenal 15 kDa... 30 1.9
TC82723 similar to GP|9294038|dbj|BAB01995.1 gene_id:MOB24.1~unk... 30 1.9
BE998760 30 2.5
AW584704 similar to SP|P05790|FBOH Fibroin heavy chain precursor... 30 2.5
BG451937 similar to GP|9758872|dbj| contains similarity to salt-... 30 2.5
BF641220 weakly similar to PIR|G86203|G86 probable N-arginine di... 30 3.3
TC81629 similar to GP|15081654|gb|AAK82482.1 AT4g26750/F10M23_90... 30 3.3
TC77995 similar to GP|21104733|dbj|BAB93321. putative receptor p... 30 3.3
TC81307 weakly similar to GP|4335772|gb|AAD17449.1| unknown prot... 30 3.3
TC78700 similar to GP|21554835|gb|AAM63703.1 unknown {Arabidopsi... 30 3.3
BQ144447 GP|12697999|d KIAA1727 protein {Homo sapiens}, partial ... 29 4.3
TC88431 glutathione synthetase GSHS1 [Medicago truncatula] 29 4.3
AJ501554 weakly similar to GP|3947733|emb| NL25 {Solanum tuberos... 29 4.3
TC85117 similar to GP|21593254|gb|AAM65203.1 S-adenosyl-L-methio... 29 4.3
>TC88781 similar to GP|9369366|gb|AAF87115.1| F10A5.18 {Arabidopsis
thaliana}, partial (67%)
Length = 1391
Score = 528 bits (1360), Expect = e-150
Identities = 269/454 (59%), Positives = 333/454 (73%), Gaps = 2/454 (0%)
Frame = +1
Query: 11 FILSSLIPHFHILIVSSSSSSPSPSPLPSLLSSPSSNQGEWELIQPTIGISAMHMQLSHN 70
F + LIP I+I+S++ + PSP+ G+W+L+Q IGI AMHMQL HN
Sbjct: 79 FPIHLLIPL--IIIISTTFTIPSPAIAAG---------GQWQLLQNNIGIVAMHMQLLHN 225
Query: 71 NKIIIFDRTDFGPSNLPLSNGRCRMDPFDTALKIDCTAHSVLYDIATNTFRSLTVQTDTW 130
+++IIFDRTDFG SNL L NGRCR++P + A+K DCTAHS+ YD+ +NTFR L +QT+ W
Sbjct: 226 DRVIIFDRTDFGFSNLSLPNGRCRVNPQEQAVKNDCTAHSLEYDVFSNTFRPLFIQTNIW 405
Query: 131 CSSGSVLSNGTLVQTGGFNDGERRIRMFTPCFNENCDWIEFPSYLSERRWYATNQILPDN 190
CSSGSV NGTL+QTGGFNDG+R +R F PC NCDW EF + LS RRWYATN LPD
Sbjct: 406 CSSGSVNPNGTLIQTGGFNDGDRSVRTFNPC--PNCDWQEFNASLSARRWYATNHKLPDG 579
Query: 191 RIIIIGGRRQFNYEFIPKTTTSSSSSSSSSIHLSFLQETNDPS-ENNLYPFVHLLPNGNL 249
R IIIGGRRQFNYEF PK ++ ++ S L FL +TND S ENNLYPFV L +GNL
Sbjct: 580 RQIIIGGRRQFNYEFYPKKDATAKNTYS----LPFLVQTNDASAENNLYPFVFLNVDGNL 747
Query: 250 FIFANTRSILFDYKQNVVVKEFPEIPGGDPHNYPSSGSSVLLPLDENQISM-EATIMICG 308
FIFAN R+ILFDY N VVK +P IPGGDP +YPSSGS+ LLPL Q + EA +MICG
Sbjct: 748 FIFANNRAILFDYNSNKVVKTYPTIPGGDPRSYPSSGSAALLPLRNLQNPLVEAEVMICG 927
Query: 309 GAPRGSFEAAKGKNFMPALKTCGFLKVTDSNPSWIIENMPMARVMGDMLILPNGDVIIIN 368
GAP+GS+E +F+ AL TCG LK+TD NPSW++E MP RVM DM++LPNGDV++IN
Sbjct: 928 GAPKGSYEKTLQGSFIGALNTCGRLKITDPNPSWVMETMPGGRVMSDMILLPNGDVLLIN 1107
Query: 369 GAGSGTAGWENGRQPVLTPVIFRSSETKSDKRFSVMSPASRPRLYHSSAIVLRDGRVLVG 428
GA G+AGWE+GR PVL P +++ + RF + P+ PR+YHS+A++LRDGRVLV
Sbjct: 1108GAAVGSAGWESGRNPVLNPFLYKPNSLVG-SRFQLQKPSDIPRMYHSTAVLLRDGRVLVA 1284
Query: 429 GSNPHVNYNFTGVEFPTDLSLEAFSPPYLSLEFD 462
GSNPHVNYNFT + FPT+L LEAFSP YL L F+
Sbjct: 1285GSNPHVNYNFTRL-FPTELRLEAFSPSYLELGFN 1383
>BF519820 weakly similar to GP|15809876|gb AT3g53950/F5K20_250 {Arabidopsis
thaliana}, partial (34%)
Length = 574
Score = 190 bits (483), Expect = 1e-48
Identities = 105/202 (51%), Positives = 136/202 (66%), Gaps = 2/202 (0%)
Frame = +1
Query: 132 SSGSVLSNGTLVQTGGFNDGERRIRMFTPCFNE-NCDWIEFPSY-LSERRWYATNQILPD 189
SSGS L NG+L+QTGG DG ++IR+F PC + +CDW E LSE RWYATNQILPD
Sbjct: 4 SSGSFLPNGSLLQTGGDLDGFKKIRIFNPCDSTGSCDWEELNDVELSEGRWYATNQILPD 183
Query: 190 NRIIIIGGRRQFNYEFIPKTTTSSSSSSSSSIHLSFLQETNDPSENNLYPFVHLLPNGNL 249
IIIIGGR EF PK + + FLQET D +NLYP+VHLLPNG L
Sbjct: 184 GSIIIIGGRGSNTVEFYPKR-------ENGVVLFPFLQETEDTQMDNLYPYVHLLPNGKL 342
Query: 250 FIFANTRSILFDYKQNVVVKEFPEIPGGDPHNYPSSGSSVLLPLDENQISMEATIMICGG 309
F+FANT+S+++D+ +N VV+ +PE+ GG P NYPS+GSSV+L L E + S EA +++CGG
Sbjct: 343 FVFANTKSVMYDFNENRVVRVYPELTGG-PRNYPSAGSSVMLAL-EGEFS-EAVVVVCGG 513
Query: 310 APRGSFEAAKGKNFMPALKTCG 331
A G+F + +PA +CG
Sbjct: 514 AQYGAF--LERNTDIPAHGSCG 573
Score = 32.3 bits (72), Expect = 0.51
Identities = 25/85 (29%), Positives = 40/85 (46%), Gaps = 4/85 (4%)
Frame = +1
Query: 358 ILPNGDVIIINGAGSGTAGW----ENGRQPVLTPVIFRSSETKSDKRFSVMSPASRPRLY 413
ILP+G +III G GS T + ENG VL P + + +T+ D + +
Sbjct: 172 ILPDGSIIIIGGRGSNTVEFYPKRENG--VVLFPFLQETEDTQMDNLYPYVH-------- 321
Query: 414 HSSAIVLRDGRVLVGGSNPHVNYNF 438
+L +G++ V + V Y+F
Sbjct: 322 -----LLPNGKLFVFANTKSVMYDF 381
>TC93790 weakly similar to GP|9369366|gb|AAF87115.1| F10A5.18 {Arabidopsis
thaliana}, partial (11%)
Length = 574
Score = 77.0 bits (188), Expect = 2e-14
Identities = 40/78 (51%), Positives = 50/78 (63%), Gaps = 4/78 (5%)
Frame = +1
Query: 497 VSVRLLAPSFTTHSFGMNQRMVVLKLIG----VTMVNLDIYYATVVGPSTQEIAPPGYYL 552
VSV + APSF THSF MNQR++VL + V + Y V P++ +APPGYYL
Sbjct: 19 VSVTMYAPSFNTHSFSMNQRLLVLDELSSSKNVVGESTSTYQVEVAIPNSPILAPPGYYL 198
Query: 553 LFLVHAGVPSSGEWVQLM 570
LF+VH VPS G WVQL+
Sbjct: 199 LFVVHQEVPSQGVWVQLL 252
>TC85101 similar to GP|15809876|gb|AAL06866.1 AT3g53950/F5K20_250
{Arabidopsis thaliana}, partial (12%)
Length = 519
Score = 53.9 bits (128), Expect = 2e-07
Identities = 25/71 (35%), Positives = 37/71 (51%)
Frame = +2
Query: 499 VRLLAPSFTTHSFGMNQRMVVLKLIGVTMVNLDIYYATVVGPSTQEIAPPGYYLLFLVHA 558
V L + F THSF QR++ L + + + P + +APPGYY+LF V+
Sbjct: 5 VNLGSAPFATHSFSQGQRLIKLGVASAMVDGDQRWRIRCTAPPSGMVAPPGYYMLFAVNQ 184
Query: 559 GVPSSGEWVQL 569
GVPS W+ +
Sbjct: 185 GVPSVARWIHM 217
>AL382796 weakly similar to GP|20161084|dbj putative glyoxal oxidase {Oryza
sativa (japonica cultivar-group)}, partial (4%)
Length = 465
Score = 53.1 bits (126), Expect = 3e-07
Identities = 34/98 (34%), Positives = 47/98 (47%), Gaps = 12/98 (12%)
Frame = +3
Query: 372 SGTAGWEN-----------GRQPVLTPVIFRSSETKSDKRFSVMSPASRPRLYHSSAIVL 420
+G AGW+ +P P+++ E K R+S + R+YHSSA ++
Sbjct: 3 TGMAGWDKKATNKTPRLHTAGKPTRNPLLYNPHEKKGS-RWSKFTEDPIIRVYHSSATLI 179
Query: 421 RDGRVLVGGSNPHVNY-NFTGVEFPTDLSLEAFSPPYL 457
DG V V GSNP+ Y E PT+ E F PPYL
Sbjct: 180 PDGTVFVAGSNPNRYYCGMDQCEHPTEHRAEKFYPPYL 293
>AL374519 similar to GP|15809876|gb| AT3g53950/F5K20_250 {Arabidopsis
thaliana}, partial (17%)
Length = 532
Score = 46.6 bits (109), Expect = 3e-05
Identities = 20/31 (64%), Positives = 26/31 (83%)
Frame = +1
Query: 403 VMSPASRPRLYHSSAIVLRDGRVLVGGSNPH 433
V++P + PRLYHS+A +L DGRVL+ GSNPH
Sbjct: 4 VLNPGTVPRLYHSTANLLPDGRVLLAGSNPH 96
>TC89368 similar to SP|O22160|TL15_ARATH Thylakoid lumenal 15 kDa protein
chloroplast precursor (p15). [Mouse-ear cress], partial
(75%)
Length = 767
Score = 30.4 bits (67), Expect = 1.9
Identities = 19/61 (31%), Positives = 34/61 (55%), Gaps = 1/61 (1%)
Frame = +2
Query: 2 SSITLILIFFILSSLIPHFHILIVSSSSSSPSPSPLPSLL-SSPSSNQGEWELIQPTIGI 60
+++ L L + S+ PH H+L + SSS+ SP+PL L ++P N + + ++G
Sbjct: 17 TTMALFLNASLPSTSKPHHHLLRSTPSSSTSSPTPLSLFLGTNPKQNHQASQKLALSLGE 196
Query: 61 S 61
S
Sbjct: 197S 199
>TC82723 similar to GP|9294038|dbj|BAB01995.1 gene_id:MOB24.1~unknown
protein {Arabidopsis thaliana}, partial (30%)
Length = 744
Score = 30.4 bits (67), Expect = 1.9
Identities = 21/52 (40%), Positives = 29/52 (55%), Gaps = 5/52 (9%)
Frame = -1
Query: 2 SSITLILIFFILSSLIPHF-----HILIVSSSSSSPSPSPLPSLLSSPSSNQ 48
S I +L+F I S + P+F IL +SSSS + S PS SS SS++
Sbjct: 276 SIICCLLVFSIFSYIFPNF*IESAKILYEASSSSLSTSSSSPSKSSSSSSSE 121
>BE998760
Length = 776
Score = 30.0 bits (66), Expect = 2.5
Identities = 22/67 (32%), Positives = 35/67 (51%)
Frame = -2
Query: 9 IFFILSSLIPHFHILIVSSSSSSPSPSPLPSLLSSPSSNQGEWELIQPTIGISAMHMQLS 68
I + S LIP + + SSSS SP PS LP S N L++ +SA+ + +
Sbjct: 487 ILTVGSILIPPYSL---SSSSFSPCPSLLPKTTSKTPKNGRLVSLVKQVCLVSALSL-VK 320
Query: 69 HNNKIII 75
H ++++I
Sbjct: 319 HVSQVLI 299
>AW584704 similar to SP|P05790|FBOH Fibroin heavy chain precursor (Fib-H)
(H-fibroin). [Silk moth] {Bombyx mori}, partial (0%)
Length = 679
Score = 30.0 bits (66), Expect = 2.5
Identities = 13/22 (59%), Positives = 18/22 (81%)
Frame = +1
Query: 26 SSSSSSPSPSPLPSLLSSPSSN 47
SS S+SPSPSP+P+ ++P SN
Sbjct: 448 SSPSASPSPSPVPAPEAAPPSN 513
>BG451937 similar to GP|9758872|dbj| contains similarity to salt-inducible
protein~gene_id:K24G6.6 {Arabidopsis thaliana}, partial
(18%)
Length = 670
Score = 30.0 bits (66), Expect = 2.5
Identities = 13/27 (48%), Positives = 17/27 (62%)
Frame = +1
Query: 19 HFHILIVSSSSSSPSPSPLPSLLSSPS 45
H H + SS S+ P PSP PS + SP+
Sbjct: 112 HHHRSLPSSPSTQPHPSPNPSTIHSPT 192
>BF641220 weakly similar to PIR|G86203|G86 probable N-arginine dibasic
convertase [imported] - Arabidopsis thaliana, partial
(5%)
Length = 634
Score = 29.6 bits (65), Expect = 3.3
Identities = 17/34 (50%), Positives = 20/34 (58%)
Frame = -2
Query: 26 SSSSSSPSPSPLPSLLSSPSSNQGEWELIQPTIG 59
SSSSSSPS S S SS SS+ I+P+ G
Sbjct: 441 SSSSSSPSSSSSSSSSSSSSSSSSSSSSIEPSFG 340
>TC81629 similar to GP|15081654|gb|AAK82482.1 AT4g26750/F10M23_90
{Arabidopsis thaliana}, partial (19%)
Length = 824
Score = 29.6 bits (65), Expect = 3.3
Identities = 22/57 (38%), Positives = 28/57 (48%), Gaps = 2/57 (3%)
Frame = +1
Query: 29 SSSPSPSPLPSLLSSPSSNQGEWELIQPTIGISAMHMQLSHNNKIIIF--DRTDFGP 83
S P PSLLSSP S ++ QPT+G S H+ LS I F +D+ P
Sbjct: 259 SQLPFSRNSPSLLSSPFSILSKFYRKQPTVGTSKPHL-LSRARCFIFFPVSSSDYKP 426
>TC77995 similar to GP|21104733|dbj|BAB93321. putative receptor protein
kinase-like protein {Oryza sativa (japonica
cultivar-group)}, partial (35%)
Length = 2455
Score = 29.6 bits (65), Expect = 3.3
Identities = 14/28 (50%), Positives = 17/28 (60%)
Frame = +2
Query: 23 LIVSSSSSSPSPSPLPSLLSSPSSNQGE 50
+I + S SPSPSP PS SPS + E
Sbjct: 881 IIPDTPSPSPSPSPFPSPSPSPSPSSSE 964
>TC81307 weakly similar to GP|4335772|gb|AAD17449.1| unknown protein
{Arabidopsis thaliana}, partial (31%)
Length = 1171
Score = 29.6 bits (65), Expect = 3.3
Identities = 19/47 (40%), Positives = 27/47 (57%)
Frame = +2
Query: 27 SSSSSPSPSPLPSLLSSPSSNQGEWELIQPTIGISAMHMQLSHNNKI 73
+SSSSPSPSP PS +SPS + + L P+ S +H S + +
Sbjct: 536 TSSSSPSPSPSPS-SASPSPSLKSFALSPPS---SFLHSPFSSSTSV 664
>TC78700 similar to GP|21554835|gb|AAM63703.1 unknown {Arabidopsis
thaliana}, partial (5%)
Length = 687
Score = 29.6 bits (65), Expect = 3.3
Identities = 17/42 (40%), Positives = 21/42 (49%), Gaps = 4/42 (9%)
Frame = -2
Query: 11 FILSSLIPHFHILIVSSSSSSPSP----SPLPSLLSSPSSNQ 48
F S L PHFH+ ++ S + SP P PS SS S Q
Sbjct: 614 FXYSQLSPHFHLYLLLQQSHTSSPPTLSQPHPSFSSSSSYPQ 489
>BQ144447 GP|12697999|d KIAA1727 protein {Homo sapiens}, partial (1%)
Length = 1379
Score = 29.3 bits (64), Expect = 4.3
Identities = 25/90 (27%), Positives = 44/90 (48%), Gaps = 1/90 (1%)
Frame = -1
Query: 2 SSITLILIFFILSSLIPHFHILIVSSSSS-SPSPSPLPSLLSSPSSNQGEWELIQPTIGI 60
S T + I++IL + F+ + + SS SP P PLP L SS+ ++P+
Sbjct: 662 SLFTYLFIYYILLHCLYMFYFFRLGAPSSPSPGPPPLP-FLPPASSHPNRCPALRPSASF 486
Query: 61 SAMHMQLSHNNKIIIFDRTDFGPSNLPLSN 90
S + ++LS ++ G S+L L++
Sbjct: 485 SLV-LRLSPTLLSLLSPPRRVGLSSLRLAH 399
>TC88431 glutathione synthetase GSHS1 [Medicago truncatula]
Length = 1952
Score = 29.3 bits (64), Expect = 4.3
Identities = 13/22 (59%), Positives = 18/22 (81%)
Frame = +2
Query: 203 YEFIPKTTTSSSSSSSSSIHLS 224
+ F P +++SSSSSSSSSI+ S
Sbjct: 113 FSFFPSSSSSSSSSSSSSINFS 178
>AJ501554 weakly similar to GP|3947733|emb| NL25 {Solanum tuberosum}, partial
(9%)
Length = 611
Score = 29.3 bits (64), Expect = 4.3
Identities = 28/98 (28%), Positives = 44/98 (44%), Gaps = 14/98 (14%)
Frame = -1
Query: 3 SITLILIFFILSSLIPHFHILIVSSS--------SSSPSPSPLPSLLSSP------SSNQ 48
+++LI ++++L S+I + IL S S SS P + S +SP S+++
Sbjct: 590 TMSLISLYWLLVSIIRNPEILAASKSASL*RIELSSVKMPRAIQSTKASPNTPFCFSTSE 411
Query: 49 GEWELIQPTIGISAMHMQLSHNNKIIIFDRTDFGPSNL 86
G G + M L H+N I R PSNL
Sbjct: 410 GS---TS*NTGTNTCFMVLRHSNIFSISSRNHVEPSNL 306
>TC85117 similar to GP|21593254|gb|AAM65203.1
S-adenosyl-L-methionine:salicylic acid carboxyl
methyltransferase-like protein, partial (29%)
Length = 557
Score = 29.3 bits (64), Expect = 4.3
Identities = 25/98 (25%), Positives = 43/98 (43%), Gaps = 18/98 (18%)
Frame = -1
Query: 7 ILIFFILSSLIPHFHILIVSSSSSSPSPSPLPSLLSSPSSNQGE-----------WELIQ 55
++ F L L+PH H ++ + SPSP P P L+ + + WEL+
Sbjct: 338 MITFTTLMVLLPHEHPKSATTKALSPSPPPPPPSLTLSRVSLKK*IIDLACMACAWELLA 159
Query: 56 -PTIGISAMH------MQLSHNNKIIIFDRTDFGPSNL 86
+ ++ H +Q+ HNN I F R+ +N+
Sbjct: 158 *AGLSFASFHAKQLLQLQIGHNN--IFFHRSHICINNI 51
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.136 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,955,656
Number of Sequences: 36976
Number of extensions: 339607
Number of successful extensions: 3760
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 3034
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3553
length of query: 570
length of database: 9,014,727
effective HSP length: 101
effective length of query: 469
effective length of database: 5,280,151
effective search space: 2476390819
effective search space used: 2476390819
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148345.6


