

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
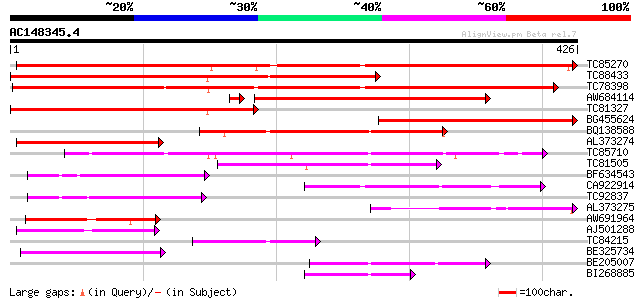
Query= AC148345.4 - phase: 0
(426 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85270 similar to GP|22854880|gb|AAN09798.1 benzoyl coenzyme A:... 476 e-135
TC88433 weakly similar to GP|22854880|gb|AAN09798.1 benzoyl coen... 446 e-126
TC78398 weakly similar to PIR|T03274|T03274 hsr201 protein hype... 425 e-119
AW684114 similar to GP|22854876|gb benzoyl coenzyme A: benzyl al... 342 2e-95
TC81327 similar to GP|22854880|gb|AAN09798.1 benzoyl coenzyme A:... 312 2e-85
BG455624 weakly similar to GP|22854876|gb benzoyl coenzyme A: be... 204 5e-53
BQ138588 similar to GP|22854876|gb| benzoyl coenzyme A: benzyl a... 176 2e-44
AL373274 similar to GP|22854880|gb benzoyl coenzyme A: benzyl al... 150 7e-37
TC85710 similar to GP|10177182|dbj|BAB10316. anthranilate N-benz... 125 2e-29
TC81505 weakly similar to GP|17104563|gb|AAL34170.1 putative N-h... 102 3e-22
BF634543 weakly similar to GP|17104563|gb putative N-hydroxycinn... 96 3e-20
CA922914 similar to GP|17104563|gb putative N-hydroxycinnamoyl/b... 96 3e-20
TC92837 similar to GP|17104563|gb|AAL34170.1 putative N-hydroxyc... 89 3e-18
AL373275 similar to GP|10140797|gb| putative hypersensitivity-re... 87 9e-18
AW691964 weakly similar to GP|6693014|gb| T22C5.6 {Arabidopsis t... 80 1e-15
AJ501288 weakly similar to GP|20160662|dbj hypothetical protein~... 79 4e-15
TC84215 weakly similar to GP|17351914|dbj|BAB78588. alcohol acet... 78 7e-15
BE325734 similar to GP|10187187|em cDNA~Lemon acyl transferase {... 71 9e-13
BE205007 similar to GP|10187187|em cDNA~Lemon acyl transferase {... 66 2e-11
BI268885 similar to GP|17104563|gb| putative N-hydroxycinnamoyl/... 62 5e-10
>TC85270 similar to GP|22854880|gb|AAN09798.1 benzoyl coenzyme A: benzyl
alcohol benzoyl transferase {Nicotiana tabacum}, partial
(94%)
Length = 1590
Score = 476 bits (1224), Expect = e-135
Identities = 247/462 (53%), Positives = 323/462 (69%), Gaps = 41/462 (8%)
Frame = +3
Query: 6 LLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVL 65
LLFTV+R PE V P+ PTP E+KLLSDIDDQE LRF +P+I Y+++P+M KDP ++
Sbjct: 30 LLFTVKRSAPEFVRPSKPTPHEIKLLSDIDDQEGLRFQIPVIQFYKYDPNMAGKDPVDII 209
Query: 66 KRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCF 125
++AL++TLV+YYP AGR+REG KLMVDCTGEGV+FIEA+ DV+L EFGDAL PPFPC
Sbjct: 210 RKALAKTLVFYYPFAGRLREGPGRKLMVDCTGEGVLFIEADADVSLKEFGDALQPPFPCL 389
Query: 126 QELIYDVPGTNQIIDHPILLIQMAR----------------------------------G 151
EL++DVPG++++++ P+L+IQ+ R G
Sbjct: 390 DELLFDVPGSSEVLNCPLLVIQVTRLKCGGFIFALRLNHTMSDAAGLVQFMSALGEISCG 569
Query: 152 AHQPSIQPVWRREILMARDPPRVTCNHHEYEQI--LSSNINKEEDVAATIVHHSFFFTLT 209
++PSI PVW RE+L +RDPPRVTC H EYEQ+ I +D+A H SFFF T
Sbjct: 570 MNEPSILPVWHRELLNSRDPPRVTCTHREYEQVPDTKGTIIPLDDMA----HRSFFFGPT 737
Query: 210 RIAEIRRLIPFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSP 269
+A+IR L+P H ++ S F+++TAC W CRT ALQ + DEEVR++CIVNAR +F + P
Sbjct: 738 EVAKIRALLPPHQQKQSNFEILTACLWRCRTIALQPDTDEEVRIICIVNARGKF---NPP 908
Query: 270 LV-GYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCL 328
L GYYGN FAFP AVTTAGKL NPLGY +EL++KAK VT+EYMHS+ADLMVIK R
Sbjct: 909 LPNGYYGNAFAFPVAVTTAGKLMENPLGYALELVKKAKPDVTQEYMHSVADLMVIKGRPH 1088
Query: 329 FTTIRSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPR-ATFIIPHKNAEGEEGLI 387
FT +RS +VSD+TRA + +V FGWG+AVYGG G +P A+F IP KNA+GEEGL+
Sbjct: 1089FTVVRSYLVSDVTRAGFGDVEFGWGKAVYGGPAKGGVGAIPGVASFYIPFKNAKGEEGLV 1268
Query: 388 LLIFLTSEVMKRFAEELDKMFGNQ-NQPTTIG--PSFVISTL 426
+ + L ++ M++F +ELD + N NQPT G PSF+IS+L
Sbjct: 1269IPLCLPAQAMEKFVKELDSVLKNNINQPTIGGPKPSFIISSL 1394
>TC88433 weakly similar to GP|22854880|gb|AAN09798.1 benzoyl coenzyme A:
benzyl alcohol benzoyl transferase {Nicotiana tabacum},
partial (57%)
Length = 990
Score = 446 bits (1148), Expect = e-126
Identities = 222/312 (71%), Positives = 243/312 (77%), Gaps = 34/312 (10%)
Frame = +3
Query: 1 MTSSPLLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKD 60
MTSSPLLFTVRRCQPELVPPAAPTP EVKLLSDIDDQE LRFN+P+IFIY HEPSM EKD
Sbjct: 57 MTSSPLLFTVRRCQPELVPPAAPTPCEVKLLSDIDDQEGLRFNIPMIFIYHHEPSMAEKD 236
Query: 61 PAKVLKRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHP 120
P KVL+ ALSQ LVYYYP AGRIREGA KLMVDCTGEGVMFIEAE DVTLD+FGDALHP
Sbjct: 237 PVKVLRHALSQALVYYYPFAGRIREGASRKLMVDCTGEGVMFIEAEADVTLDQFGDALHP 416
Query: 121 PFPCFQELIYDVPGTNQIIDHPILLIQ--------------------------------- 147
PFPCF +L+YDV G+ QI+D PI LIQ
Sbjct: 417 PFPCFHQLLYDVRGSEQIMDRPIRLIQVTRLKCGGFIVAMNWNHTMGDAAGLRQFMNAWA 596
Query: 148 -MARGAHQPSIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAATIVHHSFFF 206
MARGA++PSIQPVW REILMARDPPR+TCNH EYEQILS N KE+D A++VH SFFF
Sbjct: 597 EMARGAYRPSIQPVWNREILMARDPPRITCNHFEYEQILSPNTIKEKD-TASLVHQSFFF 773
Query: 207 TLTRIAEIRRLIPFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRAD 266
+ IA +R L+PFHLRQC+TFDLI +CFWCCRTKALQLE DEEVRMMCI NARSRF +
Sbjct: 774 RASDIATLRLLVPFHLRQCTTFDLIASCFWCCRTKALQLEADEEVRMMCIANARSRFNVN 953
Query: 267 HSPLVGYYGNCF 278
+SPLVGYY NCF
Sbjct: 954 NSPLVGYYXNCF 989
>TC78398 weakly similar to PIR|T03274|T03274 hsr201 protein
hypersensitivity-related - common tobacco, partial (75%)
Length = 1669
Score = 425 bits (1093), Expect = e-119
Identities = 230/448 (51%), Positives = 295/448 (65%), Gaps = 38/448 (8%)
Frame = +3
Query: 3 SSPLLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPA 62
S L+F VRR QPEL+ PA TP E KLLSDID Q LR N+PII YR++PSM KDP
Sbjct: 90 SPSLVFQVRRNQPELIIPATLTPHETKLLSDIDTQGGLRANVPIIQFYRNKPSMAGKDPV 269
Query: 63 KVLKRALSQTLVYYYPLAGRIREG-ARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPP 121
+V++ A++Q LV+YYPLAGR++E + KLMVDC GEGVMFIEA+ DVTL++FG+ L P
Sbjct: 270 EVIRNAIAQALVFYYPLAGRVKEADSSGKLMVDCNGEGVMFIEADADVTLEQFGE-LKAP 446
Query: 122 FPCFQELIYDVPGTNQIIDHPILLIQM--------------------------------- 148
FPC QEL+YD +++ PILLIQ+
Sbjct: 447 FPCLQELLYDAAAPEGVLNTPILLIQVTRLKCGGFIFALRFNHTMVDGVGTVHFMLAVTE 626
Query: 149 -ARGAHQPSIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAAT-IVHHSFFF 206
A+GA QP IQP W RE+L ARDPP VT NH EYEQ+ +++ + + AT SFFF
Sbjct: 627 IAKGAKQPPIQPAWHRELLNARDPPHVTFNHREYEQL--TDVQTDTVLTATEFSERSFFF 800
Query: 207 TLTRIAEIRRLIPFHLRQCST-FDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRA 265
I+ IR L+P HL ST F+++T+ W TKAL+L P EEVRMMCIV+AR +F
Sbjct: 801 GPVEISAIRNLLPRHLDNASTTFEVLTSYIWRSHTKALKLNPTEEVRMMCIVDARGKF-- 974
Query: 266 DHSPL-VGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIK 324
+ P+ VGYYGNCFAFPAAV TA ++C NPLG+ VELI+KA +V+EEYMHS+ADLMV K
Sbjct: 975 -NPPIPVGYYGNCFAFPAAVATAAEICENPLGFSVELIKKASGEVSEEYMHSVADLMVTK 1151
Query: 325 ERCLFTTIRSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPRATFIIPHKNAEGEE 384
R LFT +RSC+V D T + ++FGWG+AVYGG+ AG P F +P +NA+GEE
Sbjct: 1152GRPLFTVVRSCLVLDTTYGGFRNLDFGWGKAVYGGLAKPGAGSFPSVHFHVPGQNAKGEE 1331
Query: 385 GLILLIFLTSEVMKRFAEELDKMFGNQN 412
G+ +LI L ++VM FA+ELD + N
Sbjct: 1332GIFVLISLPTKVMTAFAKELDDLIAATN 1415
>AW684114 similar to GP|22854876|gb benzoyl coenzyme A: benzyl alcohol
benzoyl transferase {Clarkia breweri}, partial (27%)
Length = 649
Score = 342 bits (878), Expect(2) = 2e-95
Identities = 165/177 (93%), Positives = 168/177 (94%)
Frame = -3
Query: 185 LSSNINKEEDVAATIVHHSFFFTLTRIAEIRRLIPFHLRQCSTFDLITACFWCCRTKALQ 244
L SNI+KEED A TIVH SFFFTL+ IA IR LIPFHLRQCSTFD ITACFWCCRTKALQ
Sbjct: 545 LPSNISKEEDXATTIVHQSFFFTLSHIAAIRHLIPFHLRQCSTFDXITACFWCCRTKALQ 366
Query: 245 LEPDEEVRMMCIVNARSRFRADHSPLVGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRK 304
LEPDEEVRMMCIVNARSRFRADHSPLVGYYGNCFAFPAAVTTAGKLCGNPLGY VELIRK
Sbjct: 365 LEPDEEVRMMCIVNARSRFRADHSPLVGYYGNCFAFPAAVTTAGKLCGNPLGYAVELIRK 186
Query: 305 AKAQVTEEYMHSLADLMVIKERCLFTTIRSCVVSDLTRAKYAEVNFGWGEAVYGGVV 361
AKAQVTEEYMHSLADLMVIKERCLFTT+RSCVVSDLTRAKYAEVNFGWGEAVYGGVV
Sbjct: 185 AKAQVTEEYMHSLADLMVIKERCLFTTVRSCVVSDLTRAKYAEVNFGWGEAVYGGVV 15
Score = 25.0 bits (53), Expect(2) = 2e-95
Identities = 8/11 (72%), Positives = 10/11 (90%)
Frame = -2
Query: 166 LMARDPPRVTC 176
LM +DPPR+TC
Sbjct: 606 LMXKDPPRITC 574
>TC81327 similar to GP|22854880|gb|AAN09798.1 benzoyl coenzyme A: benzyl
alcohol benzoyl transferase {Nicotiana tabacum}, partial
(41%)
Length = 739
Score = 312 bits (799), Expect = 2e-85
Identities = 157/221 (71%), Positives = 168/221 (75%), Gaps = 34/221 (15%)
Frame = +3
Query: 1 MTSSPLLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKD 60
MTS+PLLFTVRR QPELVPPAAPTPREVKLLSDIDDQE LRFN+P++FIYRHEPSM EKD
Sbjct: 60 MTSAPLLFTVRRSQPELVPPAAPTPREVKLLSDIDDQEGLRFNIPMMFIYRHEPSMKEKD 239
Query: 61 PAKVLKRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHP 120
P KVL+ ALSQ LVYYYP AGRIREGA KLMVDCTGEGVMF+EAE DVTLDEFGDALHP
Sbjct: 240 PVKVLRHALSQALVYYYPFAGRIREGAGRKLMVDCTGEGVMFVEAEADVTLDEFGDALHP 419
Query: 121 PFPCFQELIYDVPGTNQIIDHPILLIQ--------------------------------- 147
P PCF+EL+YDVPG+ IID PI LIQ
Sbjct: 420 PLPCFEELLYDVPGSELIIDRPIRLIQVTRLKCGSFILVXYLNHTMSDGAGLKLFMNAWA 599
Query: 148 -MARGAHQPSIQPVWRREILMARDPPRVTCNHHEYEQILSS 187
MARGAH+PSIQPVW REILMARDPP +TCNHHEY QI SS
Sbjct: 600 EMARGAHKPSIQPVWNREILMARDPPHITCNHHEYXQIFSS 722
>BG455624 weakly similar to GP|22854876|gb benzoyl coenzyme A: benzyl alcohol
benzoyl transferase {Clarkia breweri}, partial (23%)
Length = 607
Score = 204 bits (519), Expect = 5e-53
Identities = 99/149 (66%), Positives = 115/149 (76%)
Frame = -3
Query: 278 FAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLFTTIRSCVV 337
F +PAAV AGKLCGNP GY VEL+RK KA+VTEEYMHS+AD MVIKERCLFTT+RSCV
Sbjct: 605 FXYPAAVXXAGKLCGNPFGYAVELVRKLKAEVTEEYMHSVADXMVIKERCLFTTVRSCVX 426
Query: 338 SDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPRATFIIPHKNAEGEEGLILLIFLTSEVM 397
SDLTRA+ + VNFGWGE VYGGV AG P +T+I+P+KN GEE L+L I L E M
Sbjct: 425 SDLTRARLSXVNFGWGEGVYGGVAKGGAGTFPGSTYIVPYKNINGEETLMLPICLPXEDM 246
Query: 398 KRFAEELDKMFGNQNQPTTIGPSFVISTL 426
RFA+ELD+M GNQN PT +F +STL
Sbjct: 245 XRFAKELDEMLGNQNYPTPSAHNFAMSTL 159
>BQ138588 similar to GP|22854876|gb| benzoyl coenzyme A: benzyl alcohol
benzoyl transferase {Clarkia breweri}, partial (19%)
Length = 654
Score = 176 bits (445), Expect = 2e-44
Identities = 103/198 (52%), Positives = 126/198 (63%), Gaps = 11/198 (5%)
Frame = +1
Query: 143 ILLIQMARGAHQPSIQPV---WRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAATI 199
+ L QM R S QP W REIL ARD PRVTC H EYE + N +
Sbjct: 49 VTLPQM*RFHFCSSFQPYND*WIREILSARDAPRVTCTHREYEPEPDNKANTFS--LDDM 222
Query: 200 VHHSFFFTLTRIAEIRRLIPFHLRQ-CSTFDLITACFWCCRTKALQLEPDEEVRMMCIVN 258
VHH+FFF T +A IR L+P +Q S F++IT+ FW CRT ALQL+ +EEVR++CIV+
Sbjct: 223 VHHAFFFGATEVAAIRSLLPPQQQQKYSNFEIITSYFWRCRTLALQLDTNEEVRIICIVD 402
Query: 259 ARSRFRADHSPLVGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLA 318
ARS+F P GYYGN A PAA+TT GKL NPL Y ++L++KAKA VTEEYMHS+A
Sbjct: 403 ARSKFVNLLVPN-GYYGNAIALPAAITTVGKLIENPLTYALDLVKKAKANVTEEYMHSMA 579
Query: 319 DLMVIK-------ERCLF 329
DL+VIK CLF
Sbjct: 580 DLLVIKG*TSFYYSECLF 633
>AL373274 similar to GP|22854880|gb benzoyl coenzyme A: benzyl alcohol
benzoyl transferase {Nicotiana tabacum}, partial (22%)
Length = 373
Score = 150 bits (380), Expect = 7e-37
Identities = 69/110 (62%), Positives = 87/110 (78%)
Frame = +1
Query: 6 LLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVL 65
L+F V+RC PE V P+ PTP E+KLLSDIDDQE LR+ MP+I Y + P M EKDP V+
Sbjct: 43 LVFKVKRCAPEFVRPSKPTPHEIKLLSDIDDQEGLRYQMPVIQFYNYVPVMAEKDPVDVI 222
Query: 66 KRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFG 115
++AL++TLV+YYP AGR+REG KLMVDCTGEGV+FIEA+ DVT+ + G
Sbjct: 223 RKALAKTLVFYYPFAGRLREGPGRKLMVDCTGEGVLFIEADADVTIKDLG 372
>TC85710 similar to GP|10177182|dbj|BAB10316. anthranilate
N-benzoyltransferase {Arabidopsis thaliana}, partial
(90%)
Length = 1661
Score = 125 bits (315), Expect = 2e-29
Identities = 110/408 (26%), Positives = 178/408 (42%), Gaps = 45/408 (11%)
Frame = +2
Query: 42 FNMPIIFIYRHEPSMIEKDPAKVLKRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVM 101
F+ P ++ YR + D AKVLK ALS+ LV +YP+AGR+R ++ +DC G+GV+
Sbjct: 209 FHTPSVYFYRPNGASNFFD-AKVLKEALSKVLVPFYPMAGRLRRDEDGRVEIDCDGQGVL 385
Query: 102 FIEAETDVTLDEFGDALHPPFPCFQELIYDVPGTNQIIDHPILLIQM------------- 148
F+EA+T +D+FGD P ++LI V + I +P+L++Q+
Sbjct: 386 FVEADTGGVVDDFGD--FAPTLELRQLIPAVDYSRGIETYPLLVLQVTYFKCGGVSLGVG 559
Query: 149 --------ARGAH------------QPSIQPVWRREILMARDPPRVTCNHHEYEQILSSN 188
A G H S+ P R +L ARDPPR +H EY+ S
Sbjct: 560 MQHHVADGASGLHFINTWSDVARGLDVSMPPFIDRTLLRARDPPRPVFDHIEYKPPPSMK 739
Query: 189 INKEEDVAATIVHHSFFFTLTR----IAEIRRLIPFHLRQCSTFDLITACFWCCRTKALQ 244
+++ + F LTR + + + S+++++ W KA
Sbjct: 740 THQQPTKPGSDGAAVSIFKLTREQLNTLKAKSKEAGNTIHYSSYEMLAGHVWRSVCKARS 919
Query: 245 LEPDEEVRMMCIVNARSRFRADHSPLVGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRK 304
L D+E ++ + R+R + P GY+GN + AG L P Y I
Sbjct: 920 LPDDQETKLYIATDGRARLQPPPPP--GYFGNVIFTTTPIAIAGDLMTKPTWYAASRIHN 1093
Query: 305 AKAQVTEEYMHSLADLMVIKERCLFTTIR--------SCVVSDLTRAKYAEVNFGWGEAV 356
A +++ EY+ S D + ++ L +R + ++ R + +FGWG +
Sbjct: 1094ALSRMDNEYLRSALDFLELQPD-LKALVRGAHTFKCPNLGITSWARLPIHDADFGWGRPI 1270
Query: 357 YGGVVNCVAGPLPRATFIIPHKNAEGEEGLILLIFLTSEVMKRFAEEL 404
+ G L +FIIP +G L + I L E MK F + L
Sbjct: 1271FMGPGGIAYEGL---SFIIPSSANDG--SLSVAIALQHEHMKVFKDFL 1399
>TC81505 weakly similar to GP|17104563|gb|AAL34170.1 putative
N-hydroxycinnamoyl/benzoyltransferase {Arabidopsis
thaliana}, partial (27%)
Length = 624
Score = 102 bits (254), Expect = 3e-22
Identities = 58/171 (33%), Positives = 97/171 (55%), Gaps = 3/171 (1%)
Frame = +1
Query: 157 IQPVWRREILMARDPPRVTCNHHEYEQILS-SNINKEEDVAATIVHHSFFFTLTRIAEIR 215
I P R IL +R+PP++ HHE++++ S+ NK I++ SF+F ++ ++
Sbjct: 73 IPPFLDRTILKSRNPPQIEFTHHEFDELEDLSDTNKLIHEEEEIIYKSFWFNQNKLELLK 252
Query: 216 RLIPFH--LRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPLVGY 273
+ +++CSTF+ ++A W RT+AL++ P++ V+++ V+ RSRF GY
Sbjct: 253 KKATEDGVVKKCSTFEALSAFVWRARTQALKVHPNQLVKLLFAVDGRSRFVPPIPK--GY 426
Query: 274 YGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIK 324
+GN A+ A +L NPL + VELI KA VT+ YM S+ D +K
Sbjct: 427 FGNAIVLSNAMCKARELLKNPLSFSVELINKAIEMVTDSYMRSVIDYFEVK 579
>BF634543 weakly similar to GP|17104563|gb putative
N-hydroxycinnamoyl/benzoyltransferase {Arabidopsis
thaliana}, partial (33%)
Length = 629
Score = 95.9 bits (237), Expect = 3e-20
Identities = 49/137 (35%), Positives = 83/137 (59%)
Frame = +1
Query: 14 QPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKRALSQTL 73
+P LV PA T + + LS++D + + + I+ Y+ S +D A+VLK +LS+ L
Sbjct: 142 EPTLVLPAQQTKKGLYFLSNLDQN--IAYPVRTIYCYK-SLSKGNEDVAQVLKDSLSKIL 312
Query: 74 VYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQELIYDVP 133
V+YYP+AGR++ + KL +DC EGV+F+EAE + +++ D P ++L+YD+P
Sbjct: 313 VHYYPMAGRLKISSEGKLTIDCNEEGVVFVEAEANCMVEDLSDVKEPDLDILRKLVYDIP 492
Query: 134 GTNQIIDHPILLIQMAR 150
++ P LLIQ+ +
Sbjct: 493 TAKNLLXMPPLLIQVTK 543
>CA922914 similar to GP|17104563|gb putative
N-hydroxycinnamoyl/benzoyltransferase {Arabidopsis
thaliana}, partial (36%)
Length = 756
Score = 95.5 bits (236), Expect = 3e-20
Identities = 55/182 (30%), Positives = 96/182 (52%), Gaps = 1/182 (0%)
Frame = -2
Query: 222 LRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPL-VGYYGNCFAF 280
L++CSTF+ ++A W RT AL++ P+++ +++ V+ R RF P+ GY+GN
Sbjct: 710 LKKCSTFEALSAFVWRTRTSALKMHPNQQTKLLFAVDGRPRF---IPPIPKGYFGNAIVL 540
Query: 281 PAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLFTTIRSCVVSDL 340
++ A +L NP + V L+ KA VT+ YM S D + R + + +++
Sbjct: 539 TNSLCKANELLENPFSFSVGLVHKAIDMVTDSYMRSAIDYFEV-TRARPSLTGTLLITTW 363
Query: 341 TRAKYAEVNFGWGEAVYGGVVNCVAGPLPRATFIIPHKNAEGEEGLILLIFLTSEVMKRF 400
T+ + +FGWGE + G V LP I+ + +G +G+ +L+ L + M+RF
Sbjct: 362 TKLSFDTTDFGWGEPLCSGPVT-----LPNTEVILFLSHGQGRKGISVLLGLPASAMERF 198
Query: 401 AE 402
E
Sbjct: 197 EE 192
>TC92837 similar to GP|17104563|gb|AAL34170.1 putative
N-hydroxycinnamoyl/benzoyltransferase {Arabidopsis
thaliana}, partial (28%)
Length = 605
Score = 89.0 bits (219), Expect = 3e-18
Identities = 47/135 (34%), Positives = 78/135 (56%)
Frame = +1
Query: 14 QPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKRALSQTL 73
+P LVPP+ T + + LS++D + + ++ ++++ E + +V+K AL L
Sbjct: 148 EPALVPPSEETKKGMYFLSNLDQN--IAVIVRTVYCFKNQEKGNE-NACEVIKDALKNVL 318
Query: 74 VYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQELIYDVP 133
V+YYPLAGR+ + KL+VDCTGEG +F+EAE + +++E GD P YD+P
Sbjct: 319 VHYYPLAGRLSISSEGKLIVDCTGEGALFVEAEANCSMEEIGDITKPDPRTLGMEGYDIP 498
Query: 134 GTNQIIDHPILLIQM 148
I+ P L+ Q+
Sbjct: 499 DAKHILQMPPLVAQV 543
>AL373275 similar to GP|10140797|gb| putative hypersensitivity-related
(hsr)protein {Oryza sativa}, partial (7%)
Length = 488
Score = 87.4 bits (215), Expect = 9e-18
Identities = 60/158 (37%), Positives = 81/158 (50%), Gaps = 3/158 (1%)
Frame = +3
Query: 272 GYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLFTT 331
G YGN FAFPAAVTTA +K R FT
Sbjct: 24 GNYGNSFAFPAAVTTA-----------------------------------VKSRPHFTV 98
Query: 332 IRSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPRATFIIPHKNAEGEEGLILLIF 391
+RS +V D+ A + EV+FGWG+ YGG G + +++ P KNA+G EGL++ +
Sbjct: 99 VRSYLVVDIKHAGFREVDFGWGKPCYGGPAK---GGVVYSSY-TPFKNAKGIEGLVIPVC 266
Query: 392 LTSEVMKRFAEELDKMFGNQ-NQPTTIGPS--FVISTL 426
L ++ MKRF +ELD + N+ NQPT GP F+IS L
Sbjct: 267 LPTKAMKRFIKELDGVLKNKINQPTMGGPKPRFIISPL 380
>AW691964 weakly similar to GP|6693014|gb| T22C5.6 {Arabidopsis thaliana},
partial (24%)
Length = 444
Score = 80.1 bits (196), Expect = 1e-15
Identities = 44/107 (41%), Positives = 66/107 (61%), Gaps = 6/107 (5%)
Frame = +2
Query: 13 CQP-ELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKRALSQ 71
CQP +PP+ PTP+ LS++DDQ+ LRF++ ++I++ ++ LK +LS+
Sbjct: 92 CQPITSIPPSIPTPKHSLYLSNLDDQKFLRFSIKYLYIFKKSVNLDH------LKCSLSR 253
Query: 72 TLVYYYPLAGRIREGARD-----KLMVDCTGEGVMFIEAETDVTLDE 113
LV YYPLAGR+R + + KL VDC GEG +F+EA +T E
Sbjct: 254 VLVDYYPLAGRLRSCSSNLDDEKKLEVDCNGEGALFVEAFMSITAQE 394
>AJ501288 weakly similar to GP|20160662|dbj hypothetical protein~similar to
Oryza sativa chromosome 10 OSJNBb0073N24.11, partial
(18%)
Length = 595
Score = 78.6 bits (192), Expect = 4e-15
Identities = 42/107 (39%), Positives = 64/107 (59%)
Frame = +2
Query: 6 LLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVL 65
+ +V R + +LV PA TP +S ID+ L+ N+ + ++RH P A+V+
Sbjct: 71 MAMSVTRTKRDLVKPAKETPFVTLDMSVIDNLPVLKCNVRTLHVFRHGPKA-----ARVI 235
Query: 66 KRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLD 112
+ ALS LV YYPLAGR++E L + C+G+GV ++EA T+ TLD
Sbjct: 236 REALSLALVPYYPLAGRLKESNPQCLQIKCSGDGVWYVEASTNCTLD 376
>TC84215 weakly similar to GP|17351914|dbj|BAB78588. alcohol
acetyltransferase {Cucumis melo}, partial (14%)
Length = 701
Score = 77.8 bits (190), Expect = 7e-15
Identities = 43/97 (44%), Positives = 58/97 (59%), Gaps = 1/97 (1%)
Frame = +1
Query: 138 IIDHPILLIQMARGAHQPSIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAA 197
I+ L ++++G +PSI PVW REIL ARD PRVTC H EYE + N
Sbjct: 415 IVQFMSALAEISQGMRKPSISPVWCREILSARDAPRVTCTHREYEPEPDNKANTFS--LD 588
Query: 198 TIVHHSFFFTLTRIAEIRRLIPFHLRQ-CSTFDLITA 233
+VHH+FFF T +A IR L+P +Q S F++IT+
Sbjct: 589 DMVHHAFFFGATDVAAIRSLLPPQQQQKYSNFEIITS 699
>BE325734 similar to GP|10187187|em cDNA~Lemon acyl transferase {Citrus
limon}, partial (27%)
Length = 519
Score = 70.9 bits (172), Expect = 9e-13
Identities = 44/111 (39%), Positives = 65/111 (57%), Gaps = 2/111 (1%)
Frame = +1
Query: 9 TVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKRA 68
+VR + +V P+ PTP V LS +D Q LRF + + +YR P + + LK A
Sbjct: 55 SVRVKEAVVVTPSEPTPNCVLSLSALDSQLFLRFTVEYLLVYRPGPGLDKVATTSRLKAA 234
Query: 69 LSQTLVYYYPLAGRIREGARD-KLMVDCTGEGVMFIEAETD-VTLDEFGDA 117
L++ LV YYPLAGR+R + + L V C +G +FIEA +D T+++F A
Sbjct: 235 LARALVPYYPLAGRVRPSSDNLGLEVVCKAQGAVFIEAVSDRYTVNDFEKA 387
>BE205007 similar to GP|10187187|em cDNA~Lemon acyl transferase {Citrus
limon}, partial (30%)
Length = 502
Score = 66.2 bits (160), Expect = 2e-11
Identities = 37/136 (27%), Positives = 71/136 (52%)
Frame = +1
Query: 226 STFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPLVGYYGNCFAFPAAVT 285
++F+++ A W +A+ P++ ++++ VN R+R + GYYGN F A T
Sbjct: 88 TSFEVLAAHVWRSWARAIGFPPNQTLKLLLSVNIRNRVKPGLPD--GYYGNAFVLACAQT 261
Query: 286 TAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLFTTIRSCVVSDLTRAKY 345
+A +L +GYG L++KAK +V EY+ + ++ V + R ++ +++ +R
Sbjct: 262 SAKELTEKGIGYGSGLVKKAKERVDSEYVRKVTEI-VSESRACPDSVGVLILTQWSRMGL 438
Query: 346 AEVNFGWGEAVYGGVV 361
V FG V+ G +
Sbjct: 439 ERVEFGMENPVFMGPI 486
>BI268885 similar to GP|17104563|gb| putative
N-hydroxycinnamoyl/benzoyltransferase {Arabidopsis
thaliana}, partial (15%)
Length = 266
Score = 61.6 bits (148), Expect = 5e-10
Identities = 29/84 (34%), Positives = 50/84 (59%)
Frame = +3
Query: 222 LRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPLVGYYGNCFAFP 281
L C+TF++++A W RTKAL++ P++E +++ V+ R++F GY+GN
Sbjct: 21 LESCTTFEVLSAFVWIARTKALKMLPEQETKLLFXVDGRAKFEPKLPK--GYFGNGIILT 194
Query: 282 AAVTTAGKLCGNPLGYGVELIRKA 305
AV AG++ P Y V++I+ A
Sbjct: 195 NAVCKAGEITNKPFSYTVKVIQDA 266
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.140 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,032,627
Number of Sequences: 36976
Number of extensions: 231779
Number of successful extensions: 1571
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 1518
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1541
length of query: 426
length of database: 9,014,727
effective HSP length: 99
effective length of query: 327
effective length of database: 5,354,103
effective search space: 1750791681
effective search space used: 1750791681
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148345.4


