

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148345.14 - phase: 0 /pseudo
(744 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
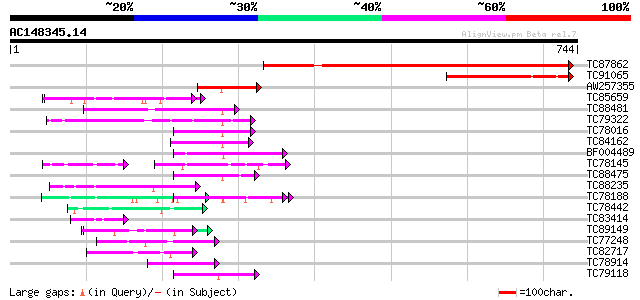
Score E
Sequences producing significant alignments: (bits) Value
TC87862 similar to GP|22022579|gb|AAM83246.1 AT3g05090/T12H1_5 {... 809 0.0
TC91065 similar to GP|22022579|gb|AAM83246.1 AT3g05090/T12H1_5 {... 272 4e-73
AW257355 similar to GP|22022579|gb| AT3g05090/T12H1_5 {Arabidops... 140 2e-33
TC85659 SP|O24076|GBLP_MEDSA Guanine nucleotide-binding protein ... 59 5e-09
TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclea... 58 2e-08
TC79322 similar to GP|13605815|gb|AAK32893.1 At2g32700/F24L7.16 ... 58 2e-08
TC78016 similar to GP|21537191|gb|AAM61532.1 PRL1 protein {Arabi... 55 1e-07
TC84162 similar to GP|15810485|gb|AAL07130.1 putative WD40-repea... 55 1e-07
BF004489 similar to GP|20161344|d contains ESTs AU086067(S4642) ... 51 2e-06
TC78145 similar to GP|15294240|gb|AAK95297.1 AT3g18860/MCB22_3 {... 51 2e-06
TC88475 similar to GP|20260442|gb|AAM13119.1 unknown protein {Ar... 50 4e-06
TC88235 similar to GP|12656803|gb|AAK00964.1 unknown protein {Or... 50 4e-06
TC78188 similar to PIR|C84870|C84870 probable splicing factor [i... 50 4e-06
TC78442 homologue to GP|5733806|gb|AAD49742.1| G protein beta su... 47 3e-05
TC83414 weakly similar to GP|17381206|gb|AAL36415.1 unknown prot... 46 5e-05
TC89149 similar to GP|20466231|gb|AAM20433.1 cell cycle switch p... 45 8e-05
TC77248 similar to GP|13489182|gb|AAK27816.1 putative WD-repeat ... 43 4e-04
TC82717 weakly similar to GP|21539583|gb|AAM53344.1 putative coa... 43 5e-04
TC78914 similar to GP|13937195|gb|AAK50091.1 At1g78070/F28K19_28... 42 7e-04
TC79118 similar to GP|16323188|gb|AAL15328.1 AT5g67320/K8K14_4 {... 42 9e-04
>TC87862 similar to GP|22022579|gb|AAM83246.1 AT3g05090/T12H1_5 {Arabidopsis
thaliana}, partial (46%)
Length = 1743
Score = 809 bits (2089), Expect = 0.0
Identities = 396/406 (97%), Positives = 396/406 (97%)
Frame = +1
Query: 334 LLCTGEHPIRQLALHNDSIWVASTDSSVHRWPAEGCDPQKIFQRGNSFLAGNLSFSRARV 393
LLCTGEHPIRQLALH DSIWVASTDSSVHRWPAEGCDPQKIFQRGNSFLAGNLSFSRARV
Sbjct: 1 LLCTGEHPIRQLALHKDSIWVASTDSSVHRWPAEGCDPQKIFQRGNSFLAGNLSFSRARV 180
Query: 394 SLEGSTPCF*IMNF**VPIYKEPTLTIRGTPGIVQHEVLNNKRHVLTKDTSDSVKLWEIT 453
SLEGSTP VPIYKEPTLTIRGTPGIVQHEVLNNKRHVLTKDTSDSVKLWEIT
Sbjct: 181 SLEGSTP---------VPIYKEPTLTIRGTPGIVQHEVLNNKRHVLTKDTSDSVKLWEIT 333
Query: 454 KGVVVEDYGKVSFKEKKEELFEMVSVPGWFTVDTRLGTLSVHLDTQHCFNAEMYSADLNI 513
KGVVVEDYGKVSFKEKKEELFEMVSVPGWFTVDTRLGTLSVHLDTQHCFNAEMYSADLNI
Sbjct: 334 KGVVVEDYGKVSFKEKKEELFEMVSVPGWFTVDTRLGTLSVHLDTQHCFNAEMYSADLNI 513
Query: 514 VGKPEDDKVNLGRETLKGLLTHWLRKRKQRMGSPAPANGELSGKDVASRSLIHSRAEVDV 573
VGKPEDDKVNLGRETLKGLLTHWLRKRKQRMGSPAPANGELSGKDVASRSLIHSRAEVDV
Sbjct: 514 VGKPEDDKVNLGRETLKGLLTHWLRKRKQRMGSPAPANGELSGKDVASRSLIHSRAEVDV 693
Query: 574 SSENDATVYPPFEFSVVSPPSIVTEGTHGGPWRKKITDLDGTEDEKDFPWWCLDCVLNNR 633
SSENDATVYPPFEFSVVSPPSIVTEGTHGGPWRKKITDLDGTEDEKDFPWWCLDCVLNNR
Sbjct: 694 SSENDATVYPPFEFSVVSPPSIVTEGTHGGPWRKKITDLDGTEDEKDFPWWCLDCVLNNR 873
Query: 634 LTPRENTKCSFYLQPCEGSSVQILTQGKLSAPRILRIHKVINYVVEKLVLDNKPLDNLHA 693
LTPRENTKCSFYLQPCEGSSVQILTQGKLSAPRILRIHKVINYVVEKLVLDNKPLDNLHA
Sbjct: 874 LTPRENTKCSFYLQPCEGSSVQILTQGKLSAPRILRIHKVINYVVEKLVLDNKPLDNLHA 1053
Query: 694 DGNFAPGVAGSQSLLPVVGDGSFRSGSGLKPWQKLRPSIEILCCNQ 739
DGNFAPGVAGSQSLLPVVGDGSFRSGSGLKPWQKLRPSIEILCCNQ
Sbjct: 1054DGNFAPGVAGSQSLLPVVGDGSFRSGSGLKPWQKLRPSIEILCCNQ 1191
>TC91065 similar to GP|22022579|gb|AAM83246.1 AT3g05090/T12H1_5 {Arabidopsis
thaliana}, partial (20%)
Length = 848
Score = 272 bits (695), Expect = 4e-73
Identities = 130/166 (78%), Positives = 146/166 (87%)
Frame = +3
Query: 574 SSENDATVYPPFEFSVVSPPSIVTEGTHGGPWRKKITDLDGTEDEKDFPWWCLDCVLNNR 633
SSEND VYPPFEFS+VSPPSI+TEGTHGG WRKKITDLDGTED KD PWWC+DCV +NR
Sbjct: 3 SSENDTMVYPPFEFSIVSPPSIITEGTHGGLWRKKITDLDGTEDGKDIPWWCVDCVSSNR 182
Query: 634 LTPRENTKCSFYLQPCEGSSVQILTQGKLSAPRILRIHKVINYVVEKLVLDNKPLDNLHA 693
L PREN+KCSFYL PCEGS+VQI TQGKLSAPRIL+I KVINYV+EKLVLD KPLD+++A
Sbjct: 183 LPPRENSKCSFYLYPCEGSNVQIHTQGKLSAPRILKIQKVINYVIEKLVLD-KPLDSVNA 359
Query: 694 DGNFAPGVAGSQSLLPVVGDGSFRSGSGLKPWQKLRPSIEILCCNQ 739
DG+F PG+AGSQ VGDGSFR SG++PWQK+RPSIEILC NQ
Sbjct: 360 DGSFPPGIAGSQLQHQTVGDGSFR--SGVRPWQKVRPSIEILCNNQ 491
>AW257355 similar to GP|22022579|gb| AT3g05090/T12H1_5 {Arabidopsis
thaliana}, partial (11%)
Length = 264
Score = 140 bits (353), Expect = 2e-33
Identities = 71/88 (80%), Positives = 74/88 (83%), Gaps = 4/88 (4%)
Frame = +1
Query: 247 TRSGSKILKLKGHTDNIRALLLDSTGRVLF----IRIIRFYDKGQQRCVHSYAVHTDSVW 302
TRSGSKILKLKGHTDN RALLLDSTGR +IR +D GQQRCVHSYAVHTDSVW
Sbjct: 1 TRSGSKILKLKGHTDNFRALLLDSTGRYCLSGSSYSMIRLWDIGQQRCVHSYAVHTDSVW 180
Query: 303 ALASTPTFSHVYSGGRDFSLYLTDLQTR 330
ALASTPT+ HVYSGG DFSLYLTDLQTR
Sbjct: 181 ALASTPTYIHVYSGGTDFSLYLTDLQTR 264
>TC85659 SP|O24076|GBLP_MEDSA Guanine nucleotide-binding protein beta
subunit-like protein. [Alfalfa] {Medicago sativa},
complete
Length = 1261
Score = 59.3 bits (142), Expect = 5e-09
Identities = 61/218 (27%), Positives = 90/218 (40%), Gaps = 7/218 (3%)
Frame = +2
Query: 46 STISDGSDYLFTGSRDGRLKRWALAEDAATCSAT---FESHVDWVNDAVLVGDNTL-VSC 101
+T D SD + T SRD + W L ++ T H +V D VL D +S
Sbjct: 125 ATPIDNSDMIVTASRDKSIILWHLTKEDKTYGVPRRRLTGHSHFVQDVVLSSDGQFALSG 304
Query: 102 SSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALA 161
S D L+ W+ L+AGT R HT V +A + N IV S + +W+
Sbjct: 305 SWDGELRLWD-LNAGTSARRFVGHTKDVLSVAFSIDNRQIV-SASRDRTIKLWNTLGECK 478
Query: 162 PVSKCNDAMVDESSNGINGSANLLPSTNLRTISSS---HSISLHTAQTQGYIPIAAKGHK 218
+ DA D S PST TI S+ ++ + T + GH
Sbjct: 479 YTIQDGDAHSDWVS-----CVRFSPSTLQPTIVSASWDRTVKVWNL-TNCKLRNTLAGHS 640
Query: 219 ESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKL 256
V +A+ GS+ SGG + V+ +WD G ++ L
Sbjct: 641 GYVNTVAVSPDGSLCASGGKDGVILLWDLAEGKRLYSL 754
Score = 51.2 bits (121), Expect = 1e-06
Identities = 54/216 (25%), Positives = 91/216 (42%), Gaps = 13/216 (6%)
Frame = +2
Query: 43 LLTSTISDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDN---TLV 99
+L+ S + + + SRD +K W + ++H DWV+ T+V
Sbjct: 383 VLSVAFSIDNRQIVSASRDRTIKLWNTLGECKYTIQDGDAHSDWVSCVRFSPSTLQPTIV 562
Query: 100 SCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAA 159
S S D T+K WN L+ T H+ YV +A + + ++ ASGG G + +WD+
Sbjct: 563 SASWDRTVKVWN-LTNCKLRNTLAGHSGYVNTVAVS-PDGSLCASGGKDGVILLWDLAEG 736
Query: 160 LAPVSKCNDAMVDE---SSN-----GINGSANLLPSTNLRTISSSHSISLHTAQTQGYIP 211
S +++ S N S+ + ++I + L T + I
Sbjct: 737 KRLYSLDAGSIIHALCFSPNRYWLCAATESSIKIWDLESKSIVEDLKVDLKT-EADAAIG 913
Query: 212 IAAKGHKESVYALAMD--EGGSILVSGGTEKVVRVW 245
K+ +Y +++ GS L SG T+ VVRVW
Sbjct: 914 GDTTTKKKVIYCTSLNWSADGSTLFSGYTDGVVRVW 1021
Score = 39.7 bits (91), Expect = 0.004
Identities = 58/324 (17%), Positives = 109/324 (32%), Gaps = 52/324 (16%)
Frame = +2
Query: 96 NTLVSCSSDTTLKTWNALSAGTCVR-THRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVW 154
+T SC+ ++ + ++ G +R T R HTD VT +A NS+++ + + +W
Sbjct: 11 HTNPSCNCNSIPQKPPIMAEGLVLRGTMRAHTDVVTAIATPIDNSDMIVTASRDKSIILW 190
Query: 155 DIEAALAPVSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAA 214
+ ++ + G+ P L
Sbjct: 191 HLTK-------------EDKTYGV-------PRRRLT----------------------- 241
Query: 215 KGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRV 274
GH V + + G +SG + +R+WD +G+ + GHT ++ ++ R
Sbjct: 242 -GHSHFVQDVVLSSDGQFALSGSWDGELRLWDLNAGTSARRFVGHTKDVLSVAFSIDNRQ 418
Query: 275 L------------------------------FIRIIRFYDKGQQRCV------------- 291
+ ++ +RF Q +
Sbjct: 419 IVSASRDRTIKLWNTLGECKYTIQDGDAHSDWVSCVRFSPSTLQPTIVSASWDRTVKVWN 598
Query: 292 -------HSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTRESSLLCTGEHPIRQ 344
++ A H+ V +A +P S SGG+D + L DL + I
Sbjct: 599 LTNCKLRNTLAGHSGYVNTVAVSPDGSLCASGGKDGVILLWDLAEGKRLYSLDAGSIIHA 778
Query: 345 LALHNDSIWV-ASTDSSVHRWPAE 367
L + W+ A+T+SS+ W E
Sbjct: 779 LCFSPNRYWLCAATESSIKIWDLE 850
>TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclear
ribonucleoprotein [imported] - Arabidopsis thaliana,
partial (60%)
Length = 1836
Score = 57.8 bits (138), Expect = 2e-08
Identities = 49/208 (23%), Positives = 88/208 (41%), Gaps = 4/208 (1%)
Frame = +1
Query: 98 LVSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIE 157
L +CS K W+ + V T + HT T +A + + N +A+ W+ +
Sbjct: 994 LATCSFTGATKLWSMPNVKK-VSTLKGHTQRATDVAYSPVHKNHLATASADRTAKYWNDQ 1170
Query: 158 AALAPVSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGH 217
AL K + + + +G L T S + L +T+ + + +GH
Sbjct: 1171 GALLGTFKGHLERLARIAFHPSGKY-------LGTASYDKTWRLWDVETEEEL-LLQEGH 1326
Query: 218 KESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFI 277
SVY L GS+ S G + + RVWD R+G +L L+GH +I + G L
Sbjct: 1327 SRSVYGLDFHHDGSLAASCGLDALARVWDLRTGRSVLALEGHVKSILGISFSPNGYHLAT 1506
Query: 278 ----RIIRFYDKGQQRCVHSYAVHTDSV 301
R +D +++ +++ H++ +
Sbjct: 1507 GGEDNTCRIWDLRKKKSLYTIPAHSNLI 1590
Score = 32.3 bits (72), Expect = 0.68
Identities = 36/194 (18%), Positives = 73/194 (37%), Gaps = 2/194 (1%)
Frame = +1
Query: 54 YLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNTLV-SCSSDTTLKTWNA 112
YL T S D + W + + E H V D +L SC D + W+
Sbjct: 1243 YLGTASYDKTWRLWDVETEEELLLQ--EGHSRSVYGLDFHHDGSLAASCGLDALARVWD- 1413
Query: 113 LSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVD 172
L G V H + ++ + N +A+GG +WD+ K
Sbjct: 1414 LRTGRSVLALEGHVKSILGISFS-PNGYHLATGGEDNTCRIWDLR-------KKKSLYTI 1569
Query: 173 ESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAA-KGHKESVYALAMDEGGS 231
+ + + P +++S+ ++ ++ + P+ GH+ V +L + G
Sbjct: 1570 PAHSNLISQVKFEPQEGYFLVTASYDMTAKVWSSRDFKPVKTLSGHEAKVTSLDVLGDGG 1749
Query: 232 ILVSGGTEKVVRVW 245
+V+ ++ +++W
Sbjct: 1750 YIVTVSHDRTIKLW 1791
Score = 28.5 bits (62), Expect = 9.8
Identities = 49/223 (21%), Positives = 79/223 (34%), Gaps = 9/223 (4%)
Frame = +1
Query: 53 DYLFTGSRDGRLKRWALAEDAATCSATFESHVDWV-NDAVLVGDNTLVSCSSDTTLKTWN 111
++L T S D K W D TF+ H++ + A L + S D T + W+
Sbjct: 1117 NHLATASADRTAKYW---NDQGALLGTFKGHLERLARIAFHPSGKYLGTASYDKTWRLWD 1287
Query: 112 ALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIE---AALAPVSKCND 168
+ + H+ V L S + AS GL VWD+ + LA
Sbjct: 1288 VETEEELL-LQEGHSRSVYGLDFHHDGS-LAASCGLDALARVWDLRTGRSVLALEGHVKS 1461
Query: 169 AM-VDESSNGIN----GSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYA 223
+ + S NG + G N +LR S ++I H+
Sbjct: 1462 ILGISFSPNGYHLATGGEDNTCRIWDLRKKKSLYTIPAHSNLIS--------------QV 1599
Query: 224 LAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRAL 266
+ G LV+ + +VW +R + L GH + +L
Sbjct: 1600 KFEPQEGYFLVTASYDMTAKVWSSRDFKPVKTLSGHEAKVTSL 1728
>TC79322 similar to GP|13605815|gb|AAK32893.1 At2g32700/F24L7.16 {Arabidopsis
thaliana}, partial (59%)
Length = 1824
Score = 57.8 bits (138), Expect = 2e-08
Identities = 62/280 (22%), Positives = 114/280 (40%), Gaps = 6/280 (2%)
Frame = +2
Query: 49 SDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNTLVSCSS-DTTL 107
SDG L + D ++ W + + +T E H + D ++T ++ SS DTT+
Sbjct: 830 SDGK-LLASAGHDKKVVLWNM--ETLKTQSTPEEHTVIITDVRFRPNSTQLATSSFDTTV 1000
Query: 108 KTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDI-EAALAPVSKC 166
+ W+A ++ + HT +V L K +++ S E+ W+I + + V K
Sbjct: 1001 RLWDAADPSVSLQAYSGHTSHVASLDFHPKKNDLFCSCDDNNEIRFWNISQYSCTRVFKG 1180
Query: 167 NDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAM 226
V S +L +S + +SL +T + + +GH V+ +
Sbjct: 1181 GSTQVRFQPR----------SGHLLAAASGNVVSLFDVETDRQMH-SLQGHSGEVHCVCW 1327
Query: 227 DEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFI----RIIRF 282
D G L S E V+VW SG I +L + + + + L + + +
Sbjct: 1328 DTNGDYLASVSQES-VKVWSLASGDCIHELNSSGNMFHSCVFHPSYSNLLVIGGYQSLEL 1504
Query: 283 YDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSL 322
+ + +C+ + H + ALA +P V S D S+
Sbjct: 1505 WHMAENKCM-TIPAHEGVISALAQSPVTGMVASASHDKSV 1621
Score = 31.6 bits (70), Expect = 1.2
Identities = 54/228 (23%), Positives = 89/228 (38%), Gaps = 34/228 (14%)
Frame = +2
Query: 52 SDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVN--DAVLVGDNTLVSCSSDTTLKT 109
S L T S D ++ W A+ + + A + H V D ++ SC + ++
Sbjct: 962 STQLATSSFDTTVRLWDAADPSVSLQA-YSGHTSHVASLDFHPKKNDLFCSCDDNNEIRF 1138
Query: 110 WNALSAGTCVRTHRQHTDYVT-------CLAAAGKN----------SNIVASGGLGGEVF 152
WN +S +C R + + V LAAA N + + G GEV
Sbjct: 1139 WN-ISQYSCTRVFKGGSTQVRFQPRSGHLLAAASGNVVSLFDVETDRQMHSLQGHSGEVH 1315
Query: 153 V--WDIEAA-LAPVSKCNDAMVDESSNG----INGSANLLPS-------TNLRTISSSHS 198
WD LA VS+ + + +S +N S N+ S +NL I S
Sbjct: 1316 CVCWDTNGDYLASVSQESVKVWSLASGDCIHELNSSGNMFHSCVFHPSYSNLLVIGGYQS 1495
Query: 199 ISL-HTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVW 245
+ L H A+ + + H+ + ALA ++ S +K V++W
Sbjct: 1496 LELWHMAENKC---MTIPAHEGVISALAQSPVTGMVASASHDKSVKIW 1630
>TC78016 similar to GP|21537191|gb|AAM61532.1 PRL1 protein {Arabidopsis
thaliana}, partial (81%)
Length = 1759
Score = 55.1 bits (131), Expect = 1e-07
Identities = 30/111 (27%), Positives = 53/111 (47%), Gaps = 4/111 (3%)
Frame = +1
Query: 216 GHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVL 275
GH V ++A+D + +G ++ +++WD SG L L GH + +R L + +
Sbjct: 580 GHLGWVRSVAVDPSNTWFATGSADRTIKIWDLASGVLKLTLTGHIEQVRGLAISHKHTYM 759
Query: 276 FI----RIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSL 322
F + ++ +D Q + + SY H V+ LA PT + +G F L
Sbjct: 760 FSAGDDKQVKCWDLEQNKVIRSYHGHLSGVYCLAIHPTIDILLTGRT*FCL 912
Score = 37.7 bits (86), Expect = 0.016
Identities = 19/68 (27%), Positives = 29/68 (41%)
Frame = +1
Query: 208 GYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALL 267
G + + GH E V LA+ + + S G +K V+ WD I GH + L
Sbjct: 682 GVLKLTLTGHIEQVRGLAISHKHTYMFSAGDDKQVKCWDLEQNKVIRSYHGHLSGVYCLA 861
Query: 268 LDSTGRVL 275
+ T +L
Sbjct: 862 IHPTIDIL 885
Score = 35.8 bits (81), Expect = 0.062
Identities = 23/87 (26%), Positives = 38/87 (43%), Gaps = 1/87 (1%)
Frame = +1
Query: 48 ISDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNT-LVSCSSDTT 106
+ + + TGS D +K W LA + T H++ V + +T + S D
Sbjct: 610 VDPSNTWFATGSADRTIKIWDLA--SGVLKLTLTGHIEQVRGLAISHKHTYMFSAGDDKQ 783
Query: 107 LKTWNALSAGTCVRTHRQHTDYVTCLA 133
+K W+ L +R++ H V CLA
Sbjct: 784 VKCWD-LEQNKVIRSYHGHLSGVYCLA 861
Score = 33.1 bits (74), Expect = 0.40
Identities = 14/54 (25%), Positives = 28/54 (50%)
Frame = +3
Query: 213 AAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRAL 266
A GH+ +V ++ +V+G + +++WD R G + L H ++RA+
Sbjct: 951 ALSGHENTVCSVFTRPTDPQVVTGSHDSTIKMWDLRYGKTMSTLTNHKKSVRAM 1112
>TC84162 similar to GP|15810485|gb|AAL07130.1 putative WD40-repeat protein
{Arabidopsis thaliana}, partial (29%)
Length = 628
Score = 54.7 bits (130), Expect = 1e-07
Identities = 28/112 (25%), Positives = 51/112 (45%), Gaps = 4/112 (3%)
Frame = +1
Query: 212 IAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDST 271
+ KGHK ++++ +V+ +K +R+W GS + +GHT ++ L +
Sbjct: 94 VVFKGHKRGIWSVEFSPVDQCVVTASGDKTIRIWAISDGSCLKTFEGHTSSVLRALFVTR 273
Query: 272 GRVLFI----RIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRD 319
G + +++ + CV +Y H D VWALA + +GG D
Sbjct: 274 GTQIVSCGADGLVKLWTVKSNECVATYDNHEDKVWALAVGRKTETLATGGSD 429
Score = 38.9 bits (89), Expect = 0.007
Identities = 34/151 (22%), Positives = 58/151 (37%)
Frame = +1
Query: 95 DNTLVSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVW 154
D +V+ S D T++ W A+S G+C++T HT V + + IV S G G V +W
Sbjct: 148 DQCVVTASGDKTIRIW-AISDGSCLKTFEGHTSSVLRALFVTRGTQIV-SCGADGLVKLW 321
Query: 155 DIEAALAPVSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAA 214
T+ S+ ++ +
Sbjct: 322 -------------------------------------TVKSNECVATY------------ 354
Query: 215 KGHKESVYALAMDEGGSILVSGGTEKVVRVW 245
H++ V+ALA+ L +GG++ VV +W
Sbjct: 355 DNHEDKVWALAVGRKTETLATGGSDAVVNLW 447
Score = 30.0 bits (66), Expect = 3.4
Identities = 27/144 (18%), Positives = 51/144 (34%), Gaps = 4/144 (2%)
Frame = +1
Query: 231 SILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFI----RIIRFYDKG 286
S++ SG ++ VW ++ KGH I ++ + + + IR +
Sbjct: 25 SLVCSGSQDRTACVWRLPDLVSVVVFKGHKRGIWSVEFSPVDQCVVTASGDKTIRIWAIS 204
Query: 287 QQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTRESSLLCTGEHPIRQLA 346
C+ ++ HT SV + + S G D + L +++ E C +
Sbjct: 205 DGSCLKTFEGHTSSVLRALFVTRGTQIVSCGADGLVKLWTVKSNE----CVATYD----- 357
Query: 347 LHNDSIWVASTDSSVHRWPAEGCD 370
H D +W + G D
Sbjct: 358 NHEDKVWALAVGRKTETLATGGSD 429
>BF004489 similar to GP|20161344|d contains ESTs AU086067(S4642)
D41819(S4642)~similar to Arabidopsis thaliana chromosome
1 At1g49040~, partial (16%)
Length = 605
Score = 50.8 bits (120), Expect = 2e-06
Identities = 40/154 (25%), Positives = 70/154 (44%), Gaps = 4/154 (2%)
Frame = +1
Query: 215 KGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRV 274
KGH +V A++ D G +VSG ++ V VWD ++ + +LKGH + + S RV
Sbjct: 16 KGHTRTVRAISSDMGK--VVSGSDDQSVLVWDKQTTQLLEELKGHDGPVSCVRTLSGERV 189
Query: 275 LFIR---IIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQ-TR 330
L ++ +D RCV + + +V + + + GRD + D++ +R
Sbjct: 190 LTASHDGTVKMWDVRTDRCVATVGRCSSAVLCMEYDDNVGILAAAGRDVVANMWDIRASR 369
Query: 331 ESSLLCTGEHPIRQLALHNDSIWVASTDSSVHRW 364
+ L IR L + D++ S D + W
Sbjct: 370 QMHKLSGHTQWIRSLRMVGDTVITGSDDWTARIW 471
Score = 41.2 bits (95), Expect = 0.001
Identities = 37/148 (25%), Positives = 65/148 (43%), Gaps = 2/148 (1%)
Frame = +1
Query: 121 THRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGING 180
T + HT V +++ + V SG V VWD + + E G +G
Sbjct: 10 TLKGHTRTVRAISS---DMGKVVSGSDDQSVLVWDKQTT----------QLLEELKGHDG 150
Query: 181 SANLLPS-TNLRTISSSHSISLHTAQTQGYIPIAAKGHKES-VYALAMDEGGSILVSGGT 238
+ + + + R +++SH ++ + +A G S V + D+ IL + G
Sbjct: 151 PVSCVRTLSGERVLTASHDGTVKMWDVRTDRCVATVGRCSSAVLCMEYDDNVGILAAAGR 330
Query: 239 EKVVRVWDTRSGSKILKLKGHTDNIRAL 266
+ V +WD R+ ++ KL GHT IR+L
Sbjct: 331 DVVANMWDIRASRQMHKLSGHTQWIRSL 414
>TC78145 similar to GP|15294240|gb|AAK95297.1 AT3g18860/MCB22_3 {Arabidopsis
thaliana}, partial (93%)
Length = 2633
Score = 50.8 bits (120), Expect = 2e-06
Identities = 53/194 (27%), Positives = 83/194 (42%), Gaps = 15/194 (7%)
Frame = +3
Query: 190 LRTISSSHSISLHTAQTQGYI-PIAAKGHKESVYALA-------MDEGGSILVSGGTEKV 241
+ T S +I L T + ++ GH V +A + GG +VSGG + +
Sbjct: 189 IATSSRDKTIRLWTRDNRKFVLDKVLVGHTSFVGPIAWIPPNPDLPHGG--VVSGGMDTL 362
Query: 242 VRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFIR--IIRFYDKGQQRCVHSYAVHTD 299
V VWD SG K+ LKGH + +++D V +R + GQ C+ ++ H
Sbjct: 363 VLVWDLNSGEKVQSLKGHQLQVTGIVVDDGDLVSSSMDCTLRRWRNGQ--CIETWEAHKG 536
Query: 300 SVWALASTPTFSHVYSGGRDFSLYL----TDLQTRESSLLCTGEHPIRQLALHND-SIWV 354
+ A+ PT + +G D +L T L T + +R LA+ +D I
Sbjct: 537 PIQAVIKLPT-GELVTGSSDTTLKTWKGKTCLHTFQGH-----ADTVRGLAVMSDLGILS 698
Query: 355 ASTDSSVHRWPAEG 368
AS D S+ W G
Sbjct: 699 ASHDGSLRLWAVSG 740
Score = 48.9 bits (115), Expect = 7e-06
Identities = 35/113 (30%), Positives = 52/113 (45%)
Frame = +3
Query: 44 LTSTISDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNTLVSCSS 103
+T + D D L + S D L+RW + C T+E+H + + + LV+ SS
Sbjct: 426 VTGIVVDDGD-LVSSSMDCTLRRWRNGQ----CIETWEAHKGPIQAVIKLPTGELVTGSS 590
Query: 104 DTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDI 156
DTTLKTW TC+ T + H D V LA + S G + +W +
Sbjct: 591 DTTLKTW---KGKTCLHTFQGHADTVRGLAVMSDLG--ILSASHDGSLRLWAV 734
Score = 40.8 bits (94), Expect = 0.002
Identities = 22/62 (35%), Positives = 29/62 (46%)
Frame = +3
Query: 55 LFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNTLVSCSSDTTLKTWNALS 114
L TGS D LK W TC TF+ H D V ++ D ++S S D +L+ W
Sbjct: 573 LVTGSSDTTLKTWK----GKTCLHTFQGHADTVRGLAVMSDLGILSASHDGSLRLWAVSG 740
Query: 115 AG 116
G
Sbjct: 741 EG 746
Score = 33.5 bits (75), Expect = 0.31
Identities = 45/185 (24%), Positives = 77/185 (41%), Gaps = 8/185 (4%)
Frame = +3
Query: 94 GDNTLVSCSS-DTTLKTWNALSAGTCV-RTHRQHTDYVTCLAAAGKNSNI----VASGGL 147
GDN ++ SS D T++ W + + + HT +V +A N ++ V SGG+
Sbjct: 174 GDNGGIATSSRDKTIRLWTRDNRKFVLDKVLVGHTSFVGPIAWIPPNPDLPHGGVVSGGM 353
Query: 148 GGEVFVWDIEAALAPVSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQ 207
V VWD+ N +S G + + +SSS +L +
Sbjct: 354 DTLVLVWDL----------NSGEKVQSLKGHQLQVTGIVVDDGDLVSSSMDCTLRRWRNG 503
Query: 208 GYIPI--AAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRA 265
I A KG ++V L E LV+G ++ ++ W + + + +GH D +R
Sbjct: 504 QCIETWEAHKGPIQAVIKLPTGE----LVTGSSDTTLKTW--KGKTCLHTFQGHADTVRG 665
Query: 266 LLLDS 270
L + S
Sbjct: 666 LAVMS 680
>TC88475 similar to GP|20260442|gb|AAM13119.1 unknown protein {Arabidopsis
thaliana}, partial (96%)
Length = 1315
Score = 49.7 bits (117), Expect = 4e-06
Identities = 33/122 (27%), Positives = 61/122 (49%), Gaps = 9/122 (7%)
Frame = +3
Query: 216 GHKESVYALAMDEGGSILVSGGTEKVVRVW--DTRSGSKI--LKLKGHTDNIRALLLDST 271
GHK+ V+++A + G+ L SG ++ R+W D + K+ ++LKGHTD++ L D
Sbjct: 195 GHKKKVHSVAWNCIGTKLASGSVDQTARIWHIDPHAHGKVKDIELKGHTDSVDQLCWDPK 374
Query: 272 GRVLFI-----RIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTD 326
L + +R +D +C + +++ + P +HV G RD L + D
Sbjct: 375 HPDLIATASGDKTVRLWDARSGKCSQQAELSGENI-NITYKPDGTHVAVGNRDDELTILD 551
Query: 327 LQ 328
++
Sbjct: 552 VR 557
>TC88235 similar to GP|12656803|gb|AAK00964.1 unknown protein {Oryza
sativa}, partial (91%)
Length = 1080
Score = 49.7 bits (117), Expect = 4e-06
Identities = 44/204 (21%), Positives = 83/204 (40%), Gaps = 6/204 (2%)
Frame = +2
Query: 53 DYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNT-LVSCSSDTTLKTWN 111
+++++GS DG +K W L A C +ES VN VL + T L+S + ++ W+
Sbjct: 482 NWMYSGSEDGTVKIWDLR--APGCQREYESRAA-VNTVVLHPNQTELISGDQNGNIRVWD 652
Query: 112 ALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMV 171
L+A +C D + ++V + G +VW + ++
Sbjct: 653 -LTANSCSCELVPEVDTAVRSLTVMWDGSLVVAANNNGTCYVWRLLRGTQTMTNFEPLHK 829
Query: 172 DESSNGINGSANLLPS-----TNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAM 226
++ NG L P L T SS H++ + + GH+ ++
Sbjct: 830 LQAHNGYILKCVLSPEFCDPHRYLATASSDHTVKIWNVD-DFTLEKTLIGHQRRLWDCVF 1006
Query: 227 DEGGSILVSGGTEKVVRVWDTRSG 250
G+ L++ ++ R+W T +G
Sbjct: 1007SVDGAYLITASSDSTARLWSTTTG 1078
Score = 33.1 bits (74), Expect = 0.40
Identities = 29/142 (20%), Positives = 56/142 (39%), Gaps = 5/142 (3%)
Frame = +2
Query: 224 LAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFIR---II 280
+ M + +L + + +R W+ +SG ++ + L + R L I
Sbjct: 206 IRMSQPSVLLATASYDHTIRFWEAKSGRCYRTIQYPDSQVNRLEITPDKRYLAAAGNPHI 385
Query: 281 RFYD--KGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTRESSLLCTG 338
R +D + V SY HT +V A+ + +YSG D ++ + DL+
Sbjct: 386 RLFDVNSNSPQPVMSYDGHTSNVMAVGFQCDGNWMYSGSEDGTVKIWDLRAPGCQREYES 565
Query: 339 EHPIRQLALHNDSIWVASTDSS 360
+ + LH + + S D +
Sbjct: 566 RAAVNTVVLHPNQTELISGDQN 631
Score = 28.5 bits (62), Expect = 9.8
Identities = 11/34 (32%), Positives = 19/34 (55%)
Frame = +2
Query: 216 GHKESVYALAMDEGGSILVSGGTEKVVRVWDTRS 249
GH +V A+ G+ + SG + V++WD R+
Sbjct: 437 GHTSNVMAVGFQCDGNWMYSGSEDGTVKIWDLRA 538
>TC78188 similar to PIR|C84870|C84870 probable splicing factor [imported] -
Arabidopsis thaliana, partial (95%)
Length = 1436
Score = 49.7 bits (117), Expect = 4e-06
Identities = 36/166 (21%), Positives = 70/166 (41%), Gaps = 8/166 (4%)
Frame = +1
Query: 215 KGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRV 274
KGHK +V L G+ ++S +K +R+WDT +G +I K+ H + + G
Sbjct: 433 KGHKNAVLDLHWTSDGTQIISASPDKTLRLWDTETGKQIKKMVEHLSYVNSCCPTRRGPP 612
Query: 275 LFIR-----IIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQT 329
L + + +D Q+ + ++ + A++ + +Y+GG D + + DL+
Sbjct: 613 LVVSGSDDGTAKLWDMRQRGSIQTFP-DKYQITAVSFSDASDKIYTGGIDNDVKIWDLRK 789
Query: 330 RESSLLCTGEHPI---RQLALHNDSIWVASTDSSVHRWPAEGCDPQ 372
E ++ G + QL+ + D + W PQ
Sbjct: 790 GEVTMTLQGHQDMITSMQLSPDGSYLLTNGMDCKLCIWDMRPYAPQ 927
Score = 44.7 bits (104), Expect = 1e-04
Identities = 38/159 (23%), Positives = 68/159 (41%), Gaps = 10/159 (6%)
Frame = +1
Query: 216 GHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSK-ILKLKGHTDNIRALLLDSTGRV 274
GH+ +VY + + GS++ SG +K + +W+ K + LKGH + + L S G
Sbjct: 307 GHQSAVYTMKFNPTGSVVASGSHDKEIFLWNVHGDCKNFMVLKGHKNAVLDLHWTSDGTQ 486
Query: 275 LFI----RIIRFYDKGQQRCVHSYAVHTDSVWALAST---PTFSHVYSGGRDFSLYLTDL 327
+ + +R +D + + H V + T P V SG D + L D+
Sbjct: 487 IISASPDKTLRLWDTETGKQIKKMVEHLSYVNSCCPTRRGPPL--VVSGSDDGTAKLWDM 660
Query: 328 QTRESSLLCTGEHPIRQLALH--NDSIWVASTDSSVHRW 364
+ R S ++ I ++ +D I+ D+ V W
Sbjct: 661 RQRGSIQTFPDKYQITAVSFSDASDKIYTGGIDNDVKIW 777
Score = 44.7 bits (104), Expect = 1e-04
Identities = 53/265 (20%), Positives = 97/265 (36%), Gaps = 43/265 (16%)
Frame = +1
Query: 42 ALLTSTISDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNT-LVS 100
A+ T + + +GS D + W + D + H + V D D T ++S
Sbjct: 319 AVYTMKFNPTGSVVASGSHDKEIFLWNVHGDCKNFMV-LKGHKNAVLDLHWTSDGTQIIS 495
Query: 101 CSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAA- 159
S D TL+ W+ G ++ +H YV + +V SG G +WD+
Sbjct: 496 ASPDKTLRLWDT-ETGKQIKKMVEHLSYVNSCCPTRRGPPLVVSGSDDGTAKLWDMRQRG 672
Query: 160 ----------LAPVS-----------------KCNDAMVDESSNGINGSANLLPSTNLRT 192
+ VS K D E + + G +++ S L
Sbjct: 673 SIQTFPDKYQITAVSFSDASDKIYTGGIDNDVKIWDLRKGEVTMTLQGHQDMITSMQLSP 852
Query: 193 -----ISSSHSISLHTAQTQGYIPI-----AAKGHK----ESVYALAMDEGGSILVSGGT 238
+++ L + Y P +GH+ +++ GS + +G +
Sbjct: 853 DGSYLLTNGMDCKLCIWDMRPYAPQNRCVKILEGHQHNFEKNLLKCGWSPDGSKVTAGSS 1032
Query: 239 EKVVRVWDTRSGSKILKLKGHTDNI 263
+++V +WDT S + KL GH ++
Sbjct: 1033DRMVYIWDTTSRRILYKLPGHNGSV 1107
>TC78442 homologue to GP|5733806|gb|AAD49742.1| G protein beta subunit
{Pisum sativum}, complete
Length = 1311
Score = 47.0 bits (110), Expect = 3e-05
Identities = 47/201 (23%), Positives = 78/201 (38%), Gaps = 17/201 (8%)
Frame = +2
Query: 76 CSATFESH------VDWVNDAVLVGDNTLVSCSSDTTLKTWNALSAGTCVRTHRQHTDYV 129
C T + H +DW ++ N +VS S D L WNAL++ +
Sbjct: 299 CCRTLQGHTGKVYSLDWTSEK-----NRIVSASQDGRLIVWNALTSQKTHAIKLPCAWVM 463
Query: 130 TCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGINGSANLLPSTN 189
TC A VA GGL ++++ + N + + G S +P +
Sbjct: 464 TC--AFSPTGQSVACGGLDSVCSIFNLNSPTDRDGNLNVSRMLSGHKGYVSSCQYVPGED 637
Query: 190 LRTISSSH---------SISLHTAQTQGYIPIAAKGHKESVYALAMDEGGS-ILVSGGTE 239
I+ S + L T+ G GH V +++++ S + VSG +
Sbjct: 638 THLITGSGDQTCVLWDITTGLRTSVFGGEFQ---SGHTADVLSISINGSNSKMFVSGSCD 808
Query: 240 KVVRVWDTRSGSKILK-LKGH 259
R+WDTR S+ ++ GH
Sbjct: 809 ATARLWDTRVASRAVRTFHGH 871
>TC83414 weakly similar to GP|17381206|gb|AAL36415.1 unknown protein
{Arabidopsis thaliana}, partial (28%)
Length = 1088
Score = 46.2 bits (108), Expect = 5e-05
Identities = 24/77 (31%), Positives = 38/77 (49%)
Frame = +2
Query: 80 FESHVDWVNDAVLVGDNTLVSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNS 139
F+ H D + D L+S S D T++ W + C+R H +YVTC+ N
Sbjct: 284 FQGHSDDILDLAWSKSGFLLSSSVDKTVRLWQ-VGISKCLRVF-SHNNYVTCVNFNPVND 457
Query: 140 NIVASGGLGGEVFVWDI 156
N SG + G+V +W++
Sbjct: 458 NFFISGSIDGKVRIWEV 508
>TC89149 similar to GP|20466231|gb|AAM20433.1 cell cycle switch protein
{Arabidopsis thaliana}, partial (71%)
Length = 1283
Score = 45.4 bits (106), Expect = 8e-05
Identities = 41/158 (25%), Positives = 65/158 (40%), Gaps = 6/158 (3%)
Frame = +2
Query: 95 DNTLVSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGE--VF 152
D L S +D L WN S +R +HT V +A + SN++ SGG + +
Sbjct: 581 DRELASGGNDNQLLVWNQHSQQPTLRL-TEHTAAVKAIAWSPHQSNLLVSGGGTADRCIR 757
Query: 153 VWDIEAALAPVSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPI 212
W+ N ++ G + NL S N+ + S+H S + Y +
Sbjct: 758 FWNTT---------NGHQLNSVDTG-SQVCNLAWSKNVNELVSTHGYSQNQIMVWKYPSL 907
Query: 213 AA----KGHKESVYALAMDEGGSILVSGGTEKVVRVWD 246
A GH V LAM G +V+G ++ +R W+
Sbjct: 908 AKVATLTGHSMRVLYLAMSPDGQTIVTGAGDETLRFWN 1021
Score = 42.7 bits (99), Expect = 5e-04
Identities = 48/182 (26%), Positives = 66/182 (35%), Gaps = 13/182 (7%)
Frame = +2
Query: 98 LVSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAG-----------KNSNIVASGG 146
LV SS TL A GTCV VT L G K + ++ G
Sbjct: 188 LVDWSSQNTL----AAGLGTCVYLWSASNSKVTKLCDLGPYDGVCSVQWTKEGSFISIGT 355
Query: 147 LGGEVFVWDIEAALAPVSKCNDAMVDESSNGINGSANLLP--STNLRTISSSHSISLHTA 204
GG+V +WD +KC + G +L S L + S +I H
Sbjct: 356 NGGQVQIWD-------GTKCKKV---RTMGGHQTRTGVLAWNSRILASGSRDRNILQHDM 505
Query: 205 QTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIR 264
+ GHK V L L SGG + + VW+ S L+L HT ++
Sbjct: 506 RVPSDFIGKLVGHKSEVCGLKWSCDDRELASGGNDNQLLVWNQHSQQPTLRLTEHTAAVK 685
Query: 265 AL 266
A+
Sbjct: 686 AI 691
>TC77248 similar to GP|13489182|gb|AAK27816.1 putative WD-repeat containing
protein {Oryza sativa}, partial (92%)
Length = 2005
Score = 43.1 bits (100), Expect = 4e-04
Identities = 36/165 (21%), Positives = 68/165 (40%), Gaps = 4/165 (2%)
Frame = +1
Query: 115 AGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDES 174
+G + T H+ VT + G+ +I+ +G V +W + + C + D S
Sbjct: 853 SGQVLATLTGHSKKVTSVKFVGQGESII-TGSADKTVRLW--QGSDDGHYNCKQILKDHS 1023
Query: 175 SN----GINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDEGG 230
+ ++ + N + +L + +S T TQ E + A G
Sbjct: 1024 AEVEAVTVHATNNYFVTASLDGSWCFYELSSGTCLTQ------VSDSSEGYTSAAFHPDG 1185
Query: 231 SILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVL 275
IL +G T+ +V++WD +S + + K GH ++ A+ G L
Sbjct: 1186 LILGTGTTDSLVKIWDVKSQANVAKFDGHVGHVTAISFSENGYYL 1320
>TC82717 weakly similar to GP|21539583|gb|AAM53344.1 putative coatomer
complex subunit {Arabidopsis thaliana}, partial (29%)
Length = 1118
Score = 42.7 bits (99), Expect = 5e-04
Identities = 36/149 (24%), Positives = 63/149 (42%), Gaps = 4/149 (2%)
Frame = +1
Query: 102 SSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALA 161
S D LK WN +C T ++ YV +A K+ + AS L G + +W I+++ A
Sbjct: 442 SDDQVLKLWNWKKGWSCDETFEGNSHYVMQVAFNPKDPSTFASASLDGTLKIWTIDSS-A 618
Query: 162 PVSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGY----IPIAAKGH 217
P + + G+N + +S S +TA+ Y +GH
Sbjct: 619 P-----NFTFEGHLKGMNCVDYFESNDKQYLLSGSDD---YTAKVWDYDSKNCVQTLEGH 774
Query: 218 KESVYALAMDEGGSILVSGGTEKVVRVWD 246
K +V A+ I+++ + V++WD
Sbjct: 775 KNNVTAICAHPEIPIIITASEDSTVKIWD 861
>TC78914 similar to GP|13937195|gb|AAK50091.1 At1g78070/F28K19_28 {Arabidopsis
thaliana}, partial (76%)
Length = 1732
Score = 42.4 bits (98), Expect = 7e-04
Identities = 28/96 (29%), Positives = 44/96 (45%), Gaps = 1/96 (1%)
Frame = +1
Query: 181 SANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEK 240
+A++ P L + + L G + KGH + ++ A G IL +G +
Sbjct: 937 NASVSPDGKLLAVLGDSTEGLIADANTGKVTGNLKGHLDYSFSSAWHPNGQILATGNQDT 1116
Query: 241 VVRVWDTRSGSK-ILKLKGHTDNIRALLLDSTGRVL 275
R+WD R+ S+ I LKG IR+L S G+ L
Sbjct: 1117 TCRLWDIRNLSESIAVLKGRMGAIRSLKFTSDGKFL 1224
Score = 28.5 bits (62), Expect = 9.8
Identities = 33/170 (19%), Positives = 66/170 (38%), Gaps = 18/170 (10%)
Frame = +1
Query: 163 VSKCNDAMVDESS--NGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKES 220
V+ C+ D+S+ N ++ N P +L+ I++++ + + + + + S
Sbjct: 757 VAFCSKITTDDSAITNAVDVFRN--PRGSLKVIAANNDSQVRVFDAENFASLGCFKYDWS 930
Query: 221 VYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFI--- 277
V ++ G +L G + D +G LKGH D + G++L
Sbjct: 931 VNNASVSPDGKLLAVLGDSTEGLIADANTGKVTGNLKGHLDYSFSSAWHPNGQILATGNQ 1110
Query: 278 -RIIRFYD-----------KGQQRCVHSYAVHTDSVW-ALASTPTFSHVY 314
R +D KG+ + S +D + A+A F H++
Sbjct: 1111DTTCRLWDIRNLSESIAVLKGRMGAIRSLKFTSDGKFLAMAEPADFVHIF 1260
>TC79118 similar to GP|16323188|gb|AAL15328.1 AT5g67320/K8K14_4 {Arabidopsis
thaliana}, partial (44%)
Length = 1098
Score = 42.0 bits (97), Expect = 9e-04
Identities = 26/126 (20%), Positives = 52/126 (40%), Gaps = 13/126 (10%)
Frame = +2
Query: 216 GHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTG--- 272
GH+ V + D G++L S + ++W + + + H+ I + TG
Sbjct: 311 GHQGEVNCVKWDPTGTLLASCSDDITAKIWSVKQDKYLHDFREHSKEIYTIRWSPTGPGT 490
Query: 273 ----------RVLFIRIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSL 322
F ++ +D + +HS H V+++A +P ++ SG D SL
Sbjct: 491 NNPNKKLLLASASFDSTVKLWDIELGKLIHSLNGHRHPVYSVAFSPNGEYIASGSLDKSL 670
Query: 323 YLTDLQ 328
++ L+
Sbjct: 671 HIWSLK 688
Score = 37.4 bits (85), Expect = 0.021
Identities = 42/189 (22%), Positives = 62/189 (32%), Gaps = 9/189 (4%)
Frame = +2
Query: 79 TFESHVDWVNDAVLVGDNTLV-SCSSDTTLKTWNALSAGTCVRTHRQHTD--YVTCLAAA 135
TF H VN TL+ SCS D T K W+ + + R+H+ Y +
Sbjct: 302 TFAGHQGEVNCVKWDPTGTLLASCSDDITAKIWS-VKQDKYLHDFREHSKEIYTIRWSPT 478
Query: 136 GKNSN------IVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGINGSANLLPSTN 189
G +N ++AS V +WDIE L+ S N
Sbjct: 479 GPGTNNPNKKLLLASASFDSTVKLWDIELG-----------------------KLIHSLN 589
Query: 190 LRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRS 249
GH+ VY++A G + SG +K + +W +
Sbjct: 590 --------------------------GHRHPVYSVAFSPNGEYIASGSLDKSLHIWSLKE 691
Query: 250 GSKILKLKG 258
G I G
Sbjct: 692 GKIIRTYNG 718
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.135 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,067,139
Number of Sequences: 36976
Number of extensions: 350217
Number of successful extensions: 1849
Number of sequences better than 10.0: 115
Number of HSP's better than 10.0 without gapping: 1677
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1811
length of query: 744
length of database: 9,014,727
effective HSP length: 103
effective length of query: 641
effective length of database: 5,206,199
effective search space: 3337173559
effective search space used: 3337173559
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148345.14


