

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148343.5 - phase: 0
(443 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
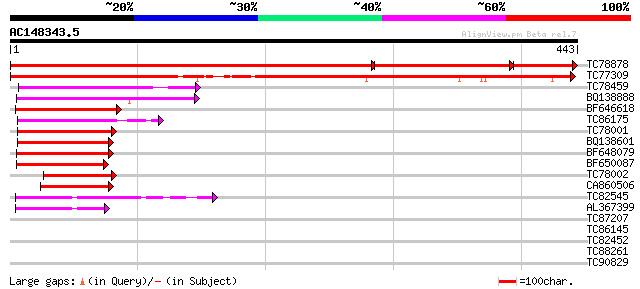
Score E
Sequences producing significant alignments: (bits) Value
TC78878 similar to GP|4519792|dbj|BAA75744.1 Asp1 {Arabidopsis t... 578 0.0
TC77309 similar to GP|4519792|dbj|BAA75744.1 Asp1 {Arabidopsis t... 610 e-175
TC78459 similar to GP|12643061|gb|AAK00450.1 unknown protein {Or... 120 1e-27
BQ138888 weakly similar to PIR|S47006|S470 zinc finger protein G... 106 2e-23
BF646618 similar to GP|21594052|gb putative GTPase activating pr... 93 2e-19
TC86175 similar to GP|23617209|dbj|BAC20880. contains ESTs C2856... 87 1e-17
TC78001 similar to GP|21618169|gb|AAM67219.1 ARF GAP-like zinc f... 87 2e-17
BQ138601 similar to GP|23617209|db contains ESTs C28562(C61610) ... 83 2e-16
BF648079 similar to GP|6648206|gb|A putative GTPase activating p... 83 2e-16
BF650087 similar to GP|6648206|gb|A putative GTPase activating p... 81 7e-16
TC78002 homologue to GP|23617209|dbj|BAC20880. contains ESTs C28... 72 6e-13
CA860506 similar to GP|12697977|dbj KIAA1716 protein {Homo sapie... 64 9e-11
TC82545 similar to GP|17064858|gb|AAL32583.1 putative protein {A... 50 2e-06
AL367399 similar to GP|17064858|gb| putative protein {Arabidopsi... 48 7e-06
TC87207 similar to GP|9802557|gb|AAF99759.1| F22O13.16 {Arabidop... 39 0.004
TC86145 homologue to GP|10334499|emb|CAC10211. hypothetical prot... 32 0.64
TC82452 weakly similar to GP|2088657|gb|AAB95289.1| unknown prot... 31 1.1
TC88261 similar to PIR|S22697|S22697 extensin - Volvox carteri (... 30 1.9
TC90829 weakly similar to PIR|T45789|T45789 hypothetical protein... 30 1.9
>TC78878 similar to GP|4519792|dbj|BAA75744.1 Asp1 {Arabidopsis thaliana},
partial (53%)
Length = 1839
Score = 578 bits (1490), Expect(3) = 0.0
Identities = 285/286 (99%), Positives = 285/286 (99%)
Frame = +2
Query: 1 MAASRRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVT 60
MAASRRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVT
Sbjct: 92 MAASRRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVT 271
Query: 61 MDSWSDLQIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSW 120
MDSWSDLQIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSW
Sbjct: 272 MDSWSDLQIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSW 451
Query: 121 RDPPVVKENASTRAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIRRNQSTGD 180
RDPPVVKENASTRAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIRRNQSTGD
Sbjct: 452 RDPPVVKENASTRAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIRRNQSTGD 631
Query: 181 VIGFVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGFFAKKMAENESRPEGLPPSQGGKY 240
VIGFVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGFFAKKMAENESRPEGLPPSQGGKY
Sbjct: 632 VIGFVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGFFAKKMAENESRPEGLPPSQGGKY 811
Query: 241 VGFGSSPGPAQRISPQNDYLSVVSEGIGKLSMVAQSATKEITAKVK 286
VGFGSSPGPAQRISPQNDYLSVVSEGIGKLSMVAQSATKEITAK K
Sbjct: 812 VGFGSSPGPAQRISPQNDYLSVVSEGIGKLSMVAQSATKEITAKGK 949
Score = 236 bits (603), Expect(3) = 0.0
Identities = 107/109 (98%), Positives = 109/109 (99%)
Frame = +1
Query: 286 KDGGYDHKVNETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNWPRN 345
+DGGYDHKVNETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNWPRN
Sbjct: 949 RDGGYDHKVNETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNWPRN 1128
Query: 346 ENDRHDFNQENKGWNSSTSTREGQPSSGGQTNTYHSNSWDDWDNQDTRK 394
ENDRHDFNQENKGWNSSTSTREGQPSSGGQTNTYHSNSWDDWDNQDTR+
Sbjct: 1129 ENDRHDFNQENKGWNSSTSTREGQPSSGGQTNTYHSNSWDDWDNQDTRE 1275
Score = 118 bits (295), Expect(3) = 0.0
Identities = 50/50 (100%), Positives = 50/50 (100%)
Frame = +2
Query: 394 KEVPAKGSAPHNNDDWAGWDDAKDDDDEFDDKSFGNNGTSGSGWTGGGFH 443
KEVPAKGSAPHNNDDWAGWDDAKDDDDEFDDKSFGNNGTSGSGWTGGGFH
Sbjct: 1274 KEVPAKGSAPHNNDDWAGWDDAKDDDDEFDDKSFGNNGTSGSGWTGGGFH 1423
>TC77309 similar to GP|4519792|dbj|BAA75744.1 Asp1 {Arabidopsis thaliana},
partial (77%)
Length = 2042
Score = 610 bits (1573), Expect = e-175
Identities = 322/495 (65%), Positives = 374/495 (75%), Gaps = 53/495 (10%)
Frame = +2
Query: 1 MAASRRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVT 60
MAASRRLR+LQS+P NKICVDCSQKNPQWASVSYG+FMCLECSGKHRGLGVHISFVRSVT
Sbjct: 155 MAASRRLRDLQSQPGNKICVDCSQKNPQWASVSYGIFMCLECSGKHRGLGVHISFVRSVT 334
Query: 61 MDSWSDLQIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSW 120
MD+WS++QIKKMEAGGN NLN FL++Y I KETDI+TKYN+NAAS+YRDRIQA++EGRSW
Sbjct: 335 MDAWSEIQIKKMEAGGNDNLNAFLAKYSIPKETDIVTKYNTNAASVYRDRIQALSEGRSW 514
Query: 121 RDPPVVKENASTRAGKGKPPLAAAS--NGGGWDDNWDNDDGDSYGYGSRGGGDIRRNQST 178
RDPPVVKEN + KG+PP+ + N GWDD WD + G GGD+ R++ST
Sbjct: 515 RDPPVVKENLGS---KGRPPMGQSRRINDSGWDD-WDREGG--------SGGDV-RSKST 655
Query: 179 GDVIGFVGNGVTPSSRSKSTEDIYTRTQLEASAANKEGFFAKKMAENESRPEGLPPSQGG 238
GD G G +RS+STEDIYTR+QLEASAA KEGFFAKKMAENESRPEGLPPSQGG
Sbjct: 656 GDFRNLNGGGA--PARSRSTEDIYTRSQLEASAAGKEGFFAKKMAENESRPEGLPPSQGG 829
Query: 239 KYVGFGSSPGPAQRISP-QNDYLSVVSEGIGKLSMVAQSA-------TKEITAKVKDGGY 290
KYVGFGSSP P+QR +P QNDY SVVS+GIGKLS+VA SA TKEIT+KVK+GGY
Sbjct: 830 KYVGFGSSPMPSQRSNPQQNDYFSVVSQGIGKLSLVAASAANAVQTGTKEITSKVKEGGY 1009
Query: 291 DHKVNETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNWPRNENDRH 350
D KVNETV +VTQKTSEIGQRTWG+MKGVMALASQKVEE+TK+ + ++NW RNE+DR+
Sbjct: 1010DQKVNETVTVVTQKTSEIGQRTWGLMKGVMALASQKVEEYTKENSNSTTNNWQRNESDRN 1189
Query: 351 ----DFNQENKGWNSSTSTRE------------------------GQP--------SSGG 374
DFNQ NKG NSST + G+P SSGG
Sbjct: 1190GYYQDFNQGNKGGNSSTGREQSSSGQYKTQSSNSWDDWGQKDSWKGEPANKGSTPYSSGG 1369
Query: 375 QTNTYHSNSWDDWDNQDTRKEVPAKGSAPHNND-DWAGWDDAKDDD-DEF-----DDKSF 427
Q+ T++ +SWDDWD++D+RKE PAK SAPHNND WAGWD+AKDD D F + K+
Sbjct: 1370QSGTHNPSSWDDWDHKDSRKEEPAKSSAPHNNDVGWAGWDNAKDDGFDNFYEGASNKKAA 1549
Query: 428 GNNGTSGSGWTGGGF 442
G+NG S WTGGGF
Sbjct: 1550GHNGKSDDAWTGGGF 1594
>TC78459 similar to GP|12643061|gb|AAK00450.1 unknown protein {Oryza
sativa}, partial (67%)
Length = 1619
Score = 120 bits (300), Expect = 1e-27
Identities = 57/142 (40%), Positives = 82/142 (57%)
Frame = +2
Query: 8 RELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSDL 67
R+L+++ NK C DC+ KNP WASV+YG+F+C++CS HR LGVHISFVRS +DSW+
Sbjct: 233 RKLKTKSENKSCFDCNAKNPTWASVTYGIFLCIDCSAVHRSLGVHISFVRSTNLDSWTPE 412
Query: 68 QIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSWRDPPVVK 127
Q+K M GGN F Q+G + + + KY S AA +Y+ + V
Sbjct: 413 QLKMMSFGGNSRAQVFFRQHGWNGDGKVEAKYTSRAAELYKQLLS-----------KEVA 559
Query: 128 ENASTRAGKGKPPLAAASNGGG 149
++ S A PP A++ + G
Sbjct: 560 KSMSEEAALSAPPAASSQSAQG 625
>BQ138888 weakly similar to PIR|S47006|S470 zinc finger protein GCS1 - yeast
(Saccharomyces cerevisiae), partial (25%)
Length = 665
Score = 106 bits (265), Expect = 2e-23
Identities = 56/151 (37%), Positives = 82/151 (54%), Gaps = 8/151 (5%)
Frame = +3
Query: 6 RLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWS 65
+L ++Q N CVDC+ +PQWAS G+FMCL CSG HRGLGVHISF+RS+TMD++
Sbjct: 114 KLLDIQKTNDNNKCVDCNAPSPQWASPKLGIFMCLSCSGIHRGLGVHISFIRSITMDAFK 293
Query: 66 DLQIKKMEAGGNRNLNTFLSQYGISKE-------TDIITKYNSNAASIYRDRIQAIAEGR 118
++ +M AGGN+ F + + + + I +Y+S A +++R+ E R
Sbjct: 294 GAELARMAAGGNKPFQEFFDAHPSNTKDNRTFEGSSIQERYDSEAGDEWKERLSCKVEDR 473
Query: 119 SWRDPPVVKE-NASTRAGKGKPPLAAASNGG 148
+ + K AG G P AS G
Sbjct: 474 EFDKSNLPKRLPKKDNAGTGAPLSGQASAAG 566
>BF646618 similar to GP|21594052|gb putative GTPase activating protein
{Arabidopsis thaliana}, partial (38%)
Length = 652
Score = 92.8 bits (229), Expect = 2e-19
Identities = 41/84 (48%), Positives = 57/84 (67%), Gaps = 1/84 (1%)
Frame = +1
Query: 5 RRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSW 64
RRL++L + N++C DC+ +P+WAS + GVF+CL+C G HR LG HIS V SVT+D W
Sbjct: 250 RRLKDLLHQKDNRVCSDCNAPDPKWASANIGVFICLKCCGVHRSLGTHISKVLSVTLDDW 429
Query: 65 SDLQIKKM-EAGGNRNLNTFLSQY 87
SD ++ M E GGN + N+ Y
Sbjct: 430 SDDEVDAMIEVGGNASANSIYEAY 501
>TC86175 similar to GP|23617209|dbj|BAC20880. contains ESTs C28562(C61610)
C25917(C11090)~similar to ARF GAP-like zinc
finger-containing protein, partial (47%)
Length = 1922
Score = 87.0 bits (214), Expect = 1e-17
Identities = 44/114 (38%), Positives = 64/114 (55%)
Frame = +2
Query: 7 LRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSD 66
L L P N+ C DC K P+WASV+ G+F+C+ CSG HR LGVHIS VRS T+D+W
Sbjct: 194 LEGLLKLPENRECADCKAKGPRWASVNLGIFICMSCSGIHRSLGVHISKVRSATLDTWLP 373
Query: 67 LQIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSW 120
Q+ +++ GN N++ E ++ T Y+ + I+A E + W
Sbjct: 374 EQVAFIQSMGNERANSYW-------EAELPTNYDRVGVENF---IRAKYEDKRW 505
>TC78001 similar to GP|21618169|gb|AAM67219.1 ARF GAP-like zinc
finger-containing protein ZIGA3 {Arabidopsis thaliana},
partial (46%)
Length = 1265
Score = 86.7 bits (213), Expect = 2e-17
Identities = 37/77 (48%), Positives = 52/77 (67%)
Frame = +1
Query: 7 LRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSD 66
L L P N+ C DC K P+WASV+ G+F+C++CSG HR LGVHIS VRS T+D+W
Sbjct: 277 LEGLLKLPENRECADCKAKAPRWASVNLGIFICMQCSGIHRSLGVHISKVRSATLDTWLP 456
Query: 67 LQIKKMEAGGNRNLNTF 83
Q+ +++ GN N++
Sbjct: 457 EQVAFIQSMGNEKANSY 507
>BQ138601 similar to GP|23617209|db contains ESTs C28562(C61610)
C25917(C11090)~similar to ARF GAP-like zinc
finger-containing protein, partial (22%)
Length = 651
Score = 83.2 bits (204), Expect = 2e-16
Identities = 38/75 (50%), Positives = 49/75 (64%)
Frame = +2
Query: 7 LRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSD 66
L L P N+ C DC K P+WASV+ G+F+C++CSG HR LGVHIS VRS T+D+W
Sbjct: 191 LEGLLKLPDNRECADCWTKAPRWASVNLGIFICMQCSGIHRSLGVHISKVRSTTLDTWLP 370
Query: 67 LQIKKMEAGGNRNLN 81
Q+ M+ GN N
Sbjct: 371 DQVSYMQFMGNVKSN 415
>BF648079 similar to GP|6648206|gb|A putative GTPase activating protein
{Arabidopsis thaliana}, partial (19%)
Length = 582
Score = 82.8 bits (203), Expect = 2e-16
Identities = 37/77 (48%), Positives = 48/77 (62%), Gaps = 1/77 (1%)
Frame = +2
Query: 6 RLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWS 65
RL L + NK C DC P+W S S GVF+C++CSG HR LGVHIS V S+ +D W+
Sbjct: 320 RLDNLMRQAGNKFCADCGSSEPKWVSSSLGVFICIKCSGIHRSLGVHISKVLSLNLDDWT 499
Query: 66 DLQIKKM-EAGGNRNLN 81
D Q+ + GGN +N
Sbjct: 500 DEQVDSLVNLGGNTLIN 550
>BF650087 similar to GP|6648206|gb|A putative GTPase activating protein
{Arabidopsis thaliana}, partial (21%)
Length = 589
Score = 81.3 bits (199), Expect = 7e-16
Identities = 36/73 (49%), Positives = 46/73 (62%), Gaps = 1/73 (1%)
Frame = +3
Query: 6 RLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWS 65
RL L + NK C DC P+W S S GVF+C++CSG HR LGVHIS V S+ +D W+
Sbjct: 360 RLDNLMRQAGNKFCADCGSSEPKWVSSSLGVFICIKCSGIHRSLGVHISKVLSLNLDDWT 539
Query: 66 DLQIKKM-EAGGN 77
D Q+ + GGN
Sbjct: 540 DEQVDSLVNLGGN 578
>TC78002 homologue to GP|23617209|dbj|BAC20880. contains ESTs C28562(C61610)
C25917(C11090)~similar to ARF GAP-like zinc
finger-containing protein, partial (26%)
Length = 737
Score = 71.6 bits (174), Expect = 6e-13
Identities = 29/57 (50%), Positives = 43/57 (74%)
Frame = +3
Query: 27 PQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSDLQIKKMEAGGNRNLNTF 83
P+WASV+ G+F+C++CSG HR LGVHIS VRS T+D+W Q+ +++ GN N++
Sbjct: 468 PRWASVNLGIFICMQCSGIHRSLGVHISKVRSATLDTWLPEQVAFIQSMGNEKANSY 638
>CA860506 similar to GP|12697977|dbj KIAA1716 protein {Homo sapiens},
partial (7%)
Length = 362
Score = 64.3 bits (155), Expect = 9e-11
Identities = 27/57 (47%), Positives = 37/57 (64%)
Frame = +3
Query: 25 KNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSDLQIKKMEAGGNRNLN 81
K+PQWAS ++ +C+ECSG HR LGVH+S V+S+ +D W I+ M GN N
Sbjct: 9 KDPQWASTNFSTLLCIECSGIHRSLGVHVSKVKSLMLDKWEPESIEVMLRLGNAKAN 179
>TC82545 similar to GP|17064858|gb|AAL32583.1 putative protein {Arabidopsis
thaliana}, partial (22%)
Length = 688
Score = 50.1 bits (118), Expect = 2e-06
Identities = 36/158 (22%), Positives = 69/158 (42%)
Frame = +2
Query: 5 RRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSW 64
R +R L N+ C++C+ PQ+ ++ F+C CSG HR V+SV+M +
Sbjct: 191 RAIRTLLKLEPNRRCINCNSLGPQYVCTNFWTFVCTNCSGIHREF---THRVKSVSMAKF 361
Query: 65 SDLQIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSWRDPP 124
+ ++ ++ GGN+ + ++ N N R+ I+ + E R +
Sbjct: 362 TSQEVTALQEGGNQRAKEIYFKEWDAQRNSFPDSSNVNR---LREFIKHVYEDRRF---- 520
Query: 125 VVKENASTRAGKGKPPLAAASNGGGWDDNWDNDDGDSY 162
+ S + +GK G DD+++N ++Y
Sbjct: 521 -TGDRTSDKPPRGK--------AGDKDDSYENRRVEAY 607
>AL367399 similar to GP|17064858|gb| putative protein {Arabidopsis thaliana},
partial (15%)
Length = 461
Score = 48.1 bits (113), Expect = 7e-06
Identities = 21/74 (28%), Positives = 41/74 (55%)
Frame = +1
Query: 5 RRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSW 64
R +R L N+ C++C+ PQ+ ++ F+C +CSG HR V+S++M +
Sbjct: 208 RTIRGLLKLTPNRRCINCNSMGPQYVCTNFWTFVCTKCSGIHREF---THRVKSISMAKF 378
Query: 65 SDLQIKKMEAGGNR 78
+ ++ ++ GGN+
Sbjct: 379 TAQEVSALQEGGNQ 420
>TC87207 similar to GP|9802557|gb|AAF99759.1| F22O13.16 {Arabidopsis
thaliana}, partial (25%)
Length = 617
Score = 38.9 bits (89), Expect = 0.004
Identities = 13/38 (34%), Positives = 23/38 (60%)
Frame = +2
Query: 7 LRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSG 44
+R L P N+ C++C+ PQ+ ++ F+C+ CSG
Sbjct: 365 IRGLMKLPPNRRCINCNSLGPQYVCTTFWTFVCITCSG 478
>TC86145 homologue to GP|10334499|emb|CAC10211. hypothetical protein {Cicer
arietinum}, partial (91%)
Length = 1311
Score = 31.6 bits (70), Expect = 0.64
Identities = 36/134 (26%), Positives = 60/134 (43%), Gaps = 8/134 (5%)
Frame = +1
Query: 305 TSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNS-DNWPRN--ENDRHDFNQENKGWNS 361
T+++G G+ ++ + E +++ + NS +N P END H+ Q+ G S
Sbjct: 112 TAKVGSGGGGVK------SNNHIREEEEEHSNLNSVENLPEKIEENDEHEAEQQQTG--S 267
Query: 362 STSTREGQPSSGGQTNTYHSNSWDDWDNQDTRKEVPAKGSAPHNND-----DWAGWDDAK 416
T+T+ QPS TN ++ D + +++ A NND W G
Sbjct: 268 ETNTQ--QPSRTPFTNLSQVDA-DLALARTLQEQERAYMMLRMNNDGSDYGSWEGGSYLH 438
Query: 417 DDDDEFDDKSFGNN 430
DD D+FDD G +
Sbjct: 439 DDGDDFDDLHDGTD 480
>TC82452 weakly similar to GP|2088657|gb|AAB95289.1| unknown protein
{Arabidopsis thaliana}, partial (12%)
Length = 1702
Score = 30.8 bits (68), Expect = 1.1
Identities = 20/91 (21%), Positives = 41/91 (44%), Gaps = 7/91 (7%)
Frame = +1
Query: 347 NDRHDFNQENKGW----NSSTSTREGQPSSGGQTNTYHSNSW---DDWDNQDTRKEVPAK 399
ND + + + GW +S EG+ + ++++S W +D +D++K
Sbjct: 46 NDIQEDSTNSGGWKAWGSSKPEVHEGESTK--VQDSWNSQKWKAAEDVSQKDSQKSSAWG 219
Query: 400 GSAPHNNDDWAGWDDAKDDDDEFDDKSFGNN 430
+ P +ND+ + W KD+ + S +N
Sbjct: 220 ATKPKSNDNRSSWGQKKDEIHVMPEDSSRSN 312
>TC88261 similar to PIR|S22697|S22697 extensin - Volvox carteri (fragment),
partial (7%)
Length = 1516
Score = 30.0 bits (66), Expect = 1.9
Identities = 23/77 (29%), Positives = 29/77 (36%)
Frame = -2
Query: 116 EGRSWRDPPVVKENASTRAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIRRN 175
EGR+ D + GK + GGG D DDG G G G G +
Sbjct: 516 EGRASPDD*GTNDTGGAEDGKERGN-GCNIGGGGGGGGGDGDDGGGGGGGEEGKGGVGGG 340
Query: 176 QSTGDVIGFVGNGVTPS 192
+S GNG+ PS
Sbjct: 339 ESR-------GNGMEPS 310
>TC90829 weakly similar to PIR|T45789|T45789 hypothetical protein F26O13.220
- Arabidopsis thaliana, partial (12%)
Length = 676
Score = 30.0 bits (66), Expect = 1.9
Identities = 25/84 (29%), Positives = 32/84 (37%), Gaps = 6/84 (7%)
Frame = +2
Query: 120 WRDPPVVKENASTRAGK--GKPPLAAASNGGGWDDNWDNDDGDSYGYGSRGGGDIRRNQS 177
W D VK AG + +SN GW+DNWD D G R G Q+
Sbjct: 272 WDDNVAVKSPVVRHAGSISANGLTSRSSNKDGWEDNWD----D*LSKGKRNKGKRYYLQN 439
Query: 178 TG----DVIGFVGNGVTPSSRSKS 197
G FVG G S +++
Sbjct: 440 WGMKRWKKANFVGEGCDKCSSARA 511
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.309 0.129 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,014,380
Number of Sequences: 36976
Number of extensions: 184021
Number of successful extensions: 1139
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 1012
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1108
length of query: 443
length of database: 9,014,727
effective HSP length: 99
effective length of query: 344
effective length of database: 5,354,103
effective search space: 1841811432
effective search space used: 1841811432
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148343.5


