

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148342.7 - phase: 0
(780 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
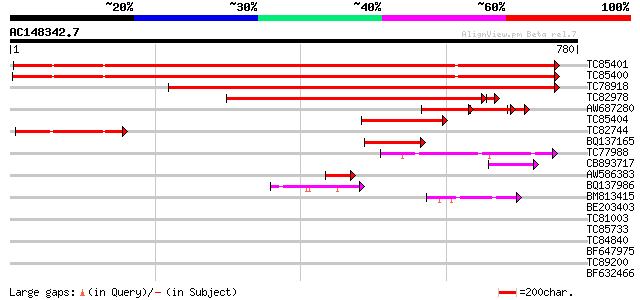
Score E
Sequences producing significant alignments: (bits) Value
TC85401 homologue to GP|13160142|emb|CAC32462. sucrose synthase ... 858 0.0
TC85400 sucrose synthase 857 0.0
TC78918 homologue to SP|O24301|SUS2_PEA Sucrose synthase 2 (EC 2... 731 0.0
TC82978 similar to PIR|C96760|C96760 probable sucrose synthase T... 603 e-176
AW687280 similar to PIR|C96760|C96 probable sucrose synthase T9L... 118 1e-51
TC85404 GP|4584690|emb|CAB40794.1 sucrose synthase {Medicago tru... 179 3e-45
TC82744 homologue to EGAD|142043|151512 second sucrose synthase ... 120 3e-27
BQ137165 similar to GP|4584690|emb| sucrose synthase {Medicago t... 104 1e-22
TC77988 homologue to GP|18375499|gb|AAK09427.2 sucrose-phosphate... 101 1e-21
CB893717 similar to SP|O04933|SPS Sucrose-phosphate synthase 2 (... 59 5e-09
AW586383 similar to GP|3377764|gb|A nodule-enhanced sucrose synt... 54 2e-07
BQ137986 homologue to SP|O04933|SPS Sucrose-phosphate synthase 2... 52 1e-06
BM813415 similar to GP|20856587|gb At1g78800/F9K20_16 {Arabidops... 44 3e-04
BE203403 similar to GP|12322785|gb unknown protein; 24439-25635 ... 39 0.010
TC81003 homologue to GP|18375499|gb|AAK09427.2 sucrose-phosphate... 36 0.049
TC85733 similar to GP|3832512|gb|AAC70779.1| granule-bound glyco... 36 0.064
TC84840 similar to PIR|C84671|C84671 NAM (no apical meristem)-li... 32 0.93
BF647975 similar to PIR|T04762|T04 chitinase homolog T16H5.170 -... 31 1.6
TC89200 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 31 2.1
BF632466 29 7.9
>TC85401 homologue to GP|13160142|emb|CAC32462. sucrose synthase isoform 3
{Pisum sativum}, complete
Length = 2846
Score = 858 bits (2218), Expect = 0.0
Identities = 421/752 (55%), Positives = 557/752 (73%), Gaps = 1/752 (0%)
Frame = +2
Query: 6 ALKRTNSIADNMPDALRKSRYHMKKCFAKYLEKGRRIMKLHELMEEVERTIDDINERNYI 65
+L R+ S+ + ++L R + ++ KG+ I++ H+++ E E ++ +R +
Sbjct: 149 SLTRSTSLRERFDESLTAHRIEILALLSRIEAKGKGILQHHQVIAEFEEIPEE--KRQKL 322
Query: 66 LEGNLGFILSSTQEAVVDPPYVAFAIRPNPGVWEYVRVNSEDLSVEPITPTDYLKFKERV 125
+ G G +L STQEAVV PP+VA A+RP PGVWEY+RV+ L V+ + +YLKFKE +
Sbjct: 323 VNGAFGEVLRSTQEAVVLPPFVALAVRPRPGVWEYLRVDVHSLVVDELRAAEYLKFKEEL 502
Query: 126 YDQKWANDENAFEADFGAFDIGIPKLTLSSSIGNGLHFVSKFLTSRTTGKLAKAQTIVDY 185
+ +N E DF F+ P+ TL+ SIGNG+ F+++ L+++ + ++++
Sbjct: 503 VEGS-SNGNFVLELDFEPFNASFPRPTLNKSIGNGVEFLNRHLSAKLFHGKESLKPLLEF 679
Query: 186 LLKLNHHGESLMINDTLSSAAKLQMALIVADVFLSAIPKDTSYQKFELRLKEWGFEKGWG 245
L N++G+++M+ND + + LQ L A+ +L I +T Y +FE + ++ G E+GWG
Sbjct: 680 LRLHNYNGKTMMVNDRIQNLDSLQHVLRTAEDYLRIIAPETPYSEFEHKFQDSGLERGWG 859
Query: 246 DNAGRVKETMRTLSEVLQAPDPVNLEIFFSRIPTIFKVVIFSVHGYFGQADVLGLPDTGG 305
D A RV E ++ L ++L+APDP LE F RIP +F VVI S HGYF Q +VLG PDTGG
Sbjct: 860 DTAERVLEMIQLLLDLLEAPDPFTLEKFLGRIPMVFNVVILSPHGYFAQDNVLGYPDTGG 1039
Query: 306 QVVYILDQVKALEEELILRIKQQGLNYKPQILVVTRLIPDARGTKCHQEFEPINDTKHSH 365
QVVYILDQV+ALE E++ RIKQQGL+ KP+IL++TRL+PDA GT C + E + DT+H H
Sbjct: 1040QVVYILDQVRALENEMLRRIKQQGLDIKPRILIITRLLPDAVGTTCGERLEKVYDTEHCH 1219
Query: 366 ILRVPFHTEKGILPQWVSRFDIYPYLERFTQDATTKILDLMEGKPDLVIGNYTDGNLVAS 425
ILRVPF TEKGI+ +W+SRF+++PYLE F++D ++ +EGKPDL++GNY+DGN+VAS
Sbjct: 1220ILRVPFRTEKGIVRKWISRFEVWPYLETFSEDVANELAKELEGKPDLIVGNYSDGNIVAS 1399
Query: 426 LMARKLGITQATIAHALEKTKYEDSDVKWKELDPKYHFSCQFMADTVAMNSSDFIITSTY 485
L+A KLG+TQ TIAHALEKTKY +S++ WK+ + KYHFSCQF AD AMN +DFIITST+
Sbjct: 1400LLAHKLGVTQCTIAHALEKTKYPESELYWKKFEDKYHFSCQFTADLFAMNHTDFIITSTF 1579
Query: 486 QEIAGSKDRPGQYESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPYTEKDQRH 545
QEIAGSKD GQYESH AFTLPGL RVV GI+VFDPKFNI +PGAD SIYFPYTE ++R
Sbjct: 1580QEIAGSKDTVGQYESHTAFTLPGLYRVVHGIDVFDPKFNIVSPGADMSIYFPYTETERRL 1759
Query: 546 SQFHPAIEDLLFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLRNL 605
+ FHP IE+LL++ V+N EHI L D+ KPIIF+MARLD VKN++GLVEWYGKN RLR L
Sbjct: 1760TSFHPEIEELLYSSVENEEHICVLKDRSKPIIFTMARLDRVKNITGLVEWYGKNARLREL 1939
Query: 606 VNLVIVGGFFDPSK-SKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNGELYRCI 664
VNLV+V G D K SKD EE AE+KKM+ LIE Y+L GQFRWI++Q +R RNGELYR I
Sbjct: 1940VNLVVVAG--DRRKESKDLEEKAEMKKMYGLIETYKLNGQFRWISSQMNRIRNGELYRVI 2113
Query: 665 ADTKGAFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLNGDESS 724
DTKGAFVQPA+YEAFGLTV+EAM CGLPTFAT GGPAEIIV G SG+HIDP +GD+++
Sbjct: 2114CDTKGAFVQPAIYEAFGLTVVEAMTCGLPTFATCNGGPAEIIVHGKSGYHIDPYHGDQAA 2293
Query: 725 NKISDFFEKCKVDPSYWNVISMAGLQRINEWY 756
+ +FFEK K DPSYW+ IS GL+RI+E Y
Sbjct: 2294ETLVEFFEKSKADPSYWDKISHGGLKRIHEKY 2389
>TC85400 sucrose synthase
Length = 3133
Score = 857 bits (2214), Expect = 0.0
Identities = 424/754 (56%), Positives = 554/754 (73%), Gaps = 1/754 (0%)
Frame = +2
Query: 4 THALKRTNSIADNMPDALRKSRYHMKKCFAKYLEKGRRIMKLHELMEEVERTIDDINERN 63
T L R +S+ + + + L +R + ++ KG+ I++ H+++ E E +D R
Sbjct: 323 TERLTRVHSLKERLDETLTANRNEILALLSRLEAKGKGILQHHQVIAEFEEIPED--SRQ 496
Query: 64 YILEGNLGFILSSTQEAVVDPPYVAFAIRPNPGVWEYVRVNSEDLSVEPITPTDYLKFKE 123
+ +G G +L STQEA+V PP+VA A+RP PG+WEY+RVN L VE + P ++LKFKE
Sbjct: 497 KLTDGAFGEVLRSTQEAIVLPPWVALAVRPRPGIWEYLRVNVHALVVENLQPAEFLKFKE 676
Query: 124 RVYDQKWANDENAFEADFGAFDIGIPKLTLSSSIGNGLHFVSKFLTSRTTGKLAKAQTIV 183
+ D AN E DF F P+ TL+ SIGNG+ F+++ L+++ ++
Sbjct: 677 ELVDGS-ANGNFVLELDFEPFTASFPRPTLNKSIGNGVQFLNRHLSAKLFHDKESLHPLL 853
Query: 184 DYLLKLNHHGESLMINDTLSSAAKLQMALIVADVFLSAIPKDTSYQKFELRLKEWGFEKG 243
++L ++ G++LM+ND + + LQ L A+ +LS I +T Y +FE R +E G E+G
Sbjct: 854 EFLRLHSYKGKTLMLNDRIQNPDSLQHVLRKAEEYLSTIDPETPYSEFEHRFQEIGLERG 1033
Query: 244 WGDNAGRVKETMRTLSEVLQAPDPVNLEIFFSRIPTIFKVVIFSVHGYFGQADVLGLPDT 303
WGD A RV E+++ L ++L+APDP LE F RIP +F VVI S HGYF Q DVLG PDT
Sbjct: 1034 WGDTAERVLESIQLLLDLLEAPDPCTLETFLDRIPMVFNVVILSPHGYFAQDDVLGYPDT 1213
Query: 304 GGQVVYILDQVKALEEELILRIKQQGLNYKPQILVVTRLIPDARGTKCHQEFEPINDTKH 363
GGQVVYILDQV+ALE E++ RIK+QGL+ P+IL++TRL+PDA GT C Q E + T+H
Sbjct: 1214 GGQVVYILDQVRALESEMLSRIKKQGLDIIPRILIITRLLPDAVGTTCGQRLEKVYGTEH 1393
Query: 364 SHILRVPFHTEKGILPQWVSRFDIYPYLERFTQDATTKILDLMEGKPDLVIGNYTDGNLV 423
HILRVPF KGI+ +W+SRF+++PYLE +T+D ++ ++GKPDL++GNY+DGN+V
Sbjct: 1394 CHILRVPFRDTKGIVRKWISRFEVWPYLETYTEDVAHELAKELQGKPDLIVGNYSDGNIV 1573
Query: 424 ASLMARKLGITQATIAHALEKTKYEDSDVKWKELDPKYHFSCQFMADTVAMNSSDFIITS 483
ASL+A KLG+TQ TIAHALEKTKY +SD+ WK+ + KYHFSCQF AD AMN +DFIITS
Sbjct: 1574 ASLLAHKLGVTQCTIAHALEKTKYPESDIYWKKFEEKYHFSCQFTADLFAMNHTDFIITS 1753
Query: 484 TYQEIAGSKDRPGQYESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPYTEKDQ 543
T+QEIAGSKD+ GQYESH AFTLPGL RVV GI+VFDPKFNI +PGADQ+IYFPYTE +
Sbjct: 1754 TFQEIAGSKDKVGQYESHTAFTLPGLYRVVHGIDVFDPKFNIVSPGADQTIYFPYTETSR 1933
Query: 544 RHSQFHPAIEDLLFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLR 603
R + F+P IE+LL++ V+N EHI L D+ KPIIF+MARLD VKN++GLVEWYGKN +LR
Sbjct: 1934 RLTSFYPEIEELLYSSVENEEHICVLKDRNKPIIFTMARLDRVKNITGLVEWYGKNAKLR 2113
Query: 604 NLVNLVIVGGFFDPSK-SKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNGELYR 662
LVNLV+V G D K SKD EE+AE+KKM+ LIE Y+L GQFRWI++Q +R RNGELYR
Sbjct: 2114 ELVNLVVVAG--DRRKESKDLEEIAEMKKMYGLIETYKLNGQFRWISSQMNRVRNGELYR 2287
Query: 663 CIADTKGAFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLNGDE 722
I DTKGAFVQPA+YEAFGLTV+EAM GLPTFAT GGPAEIIV G SGFHIDP +GD
Sbjct: 2288 VICDTKGAFVQPAVYEAFGLTVVEAMATGLPTFATLNGGPAEIIVHGKSGFHIDPYHGDR 2467
Query: 723 SSNKISDFFEKCKVDPSYWNVISMAGLQRINEWY 756
+++ + +FFEK KVDPS+W+ IS GLQRI E Y
Sbjct: 2468 AADLLVEFFEKVKVDPSHWDKISQGGLQRIEEKY 2569
>TC78918 homologue to SP|O24301|SUS2_PEA Sucrose synthase 2 (EC 2.4.1.13)
(Sucrose-UDP glucosyltransferase 2). [Garden pea] {Pisum
sativum}, partial (73%)
Length = 1985
Score = 731 bits (1886), Expect = 0.0
Identities = 350/538 (65%), Positives = 434/538 (80%)
Frame = +2
Query: 219 LSAIPKDTSYQKFELRLKEWGFEKGWGDNAGRVKETMRTLSEVLQAPDPVNLEIFFSRIP 278
LS + DT Y +FE L+ GFE+GWGD A RV ETM L ++LQAPDP LE F R+P
Sbjct: 14 LSKLAPDTLYSEFEYVLQGMGFERGWGDTAARVLETMHLLLDILQAPDPSTLETFLGRVP 193
Query: 279 TIFKVVIFSVHGYFGQADVLGLPDTGGQVVYILDQVKALEEELILRIKQQGLNYKPQILV 338
+F VVI S HG+FGQA+VLGLPDTGGQVVYILDQV+ALE E++ RI++QGL++ P+IL+
Sbjct: 194 MVFNVVILSPHGFFGQANVLGLPDTGGQVVYILDQVRALENEMLARIQKQGLDFTPRILI 373
Query: 339 VTRLIPDARGTKCHQEFEPINDTKHSHILRVPFHTEKGILPQWVSRFDIYPYLERFTQDA 398
VTRLIPDA+GT C+Q E ++ T ++HILRVPF +EKGIL +W+SRFD++P+LE + +D
Sbjct: 374 VTRLIPDAKGTTCNQRLERVSGTDYTHILRVPFRSEKGILRKWISRFDVWPFLETYAEDV 553
Query: 399 TTKILDLMEGKPDLVIGNYTDGNLVASLMARKLGITQATIAHALEKTKYEDSDVKWKELD 458
++I ++ PD +IGNY+DGNLVASL+A K+G+TQ TIAHALEKTKY DSD+ WK+ +
Sbjct: 554 ASEISAELQCYPDFIIGNYSDGNLVASLLAYKMGVTQCTIAHALEKTKYPDSDIYWKKFE 733
Query: 459 PKYHFSCQFMADTVAMNSSDFIITSTYQEIAGSKDRPGQYESHAAFTLPGLCRVVSGINV 518
KYHFSCQF AD +AMN++DFIITSTYQEIAG+K+ GQYESH AFTLPGL RVV GI+V
Sbjct: 734 DKYHFSCQFTADLIAMNNADFIITSTYQEIAGTKNTVGQYESHTAFTLPGLYRVVHGIDV 913
Query: 519 FDPKFNIAAPGADQSIYFPYTEKDQRHSQFHPAIEDLLFNKVDNNEHIGYLADKRKPIIF 578
FDPKFNI +PGAD +IYFP +EK++R + H +IE LL++ +E+IG LAD+ KPIIF
Sbjct: 914 FDPKFNIVSPGADMTIYFPCSEKEKRLTALHGSIEKLLYDTEQTDEYIGSLADRSKPIIF 1093
Query: 579 SMARLDVVKNLSGLVEWYGKNKRLRNLVNLVIVGGFFDPSKSKDREEMAEIKKMHDLIEK 638
SMARLD VKN++GLVE Y KN +LR LVNLV+V G+ D KS DREE+AEI+KMHDL+++
Sbjct: 1094SMARLDRVKNITGLVESYAKNSKLRELVNLVVVAGYIDVKKSSDREEIAEIEKMHDLMKQ 1273
Query: 639 YQLKGQFRWIAAQTDRYRNGELYRCIADTKGAFVQPALYEAFGLTVIEAMNCGLPTFATN 698
Y L G+FRWI AQT+R RNGELYR IADTKGAFVQPA YEAFGLTV+EAM CGLPTFAT
Sbjct: 1274YNLNGEFRWITAQTNRARNGELYRYIADTKGAFVQPAFYEAFGLTVVEAMTCGLPTFATC 1453
Query: 699 QGGPAEIIVDGVSGFHIDPLNGDESSNKISDFFEKCKVDPSYWNVISMAGLQRINEWY 756
GGPAEII GVSGFHIDP + D++S + +FF+KCK DP++WN IS GLQRI E Y
Sbjct: 1454HGGPAEIIEHGVSGFHIDPYHPDKASELLLEFFQKCKEDPNHWNKISDGGLQRIFERY 1627
>TC82978 similar to PIR|C96760|C96760 probable sucrose synthase T9L24.42
[imported] - Arabidopsis thaliana, partial (39%)
Length = 1127
Score = 603 bits (1554), Expect(2) = e-176
Identities = 291/358 (81%), Positives = 326/358 (90%)
Frame = +3
Query: 299 GLPDTGGQVVYILDQVKALEEELILRIKQQGLNYKPQILVVTRLIPDARGTKCHQEFEPI 358
GLPDTGGQVVYILDQV+ALEEEL+ +I+ QGLN KPQILVVTRLIP+A+GT C+QE EPI
Sbjct: 3 GLPDTGGQVVYILDQVRALEEELLQKIELQGLNVKPQILVVTRLIPNAKGTTCNQELEPI 182
Query: 359 NDTKHSHILRVPFHTEKGILPQWVSRFDIYPYLERFTQDATTKILDLMEGKPDLVIGNYT 418
TKHSHILRVPF TEKGIL QWVSRFDIYPYLERF QD+T KIL+LM+GKPDL+IGNYT
Sbjct: 183 IKTKHSHILRVPFWTEKGILSQWVSRFDIYPYLERFAQDSTIKILELMDGKPDLIIGNYT 362
Query: 419 DGNLVASLMARKLGITQATIAHALEKTKYEDSDVKWKELDPKYHFSCQFMADTVAMNSSD 478
DGNLV+SLMA KLG+TQATIAHALEKTKYEDSD KW + KYHFS QF AD +AMNS+D
Sbjct: 363 DGNLVSSLMASKLGVTQATIAHALEKTKYEDSDAKWNSFEEKYHFSSQFTADLIAMNSAD 542
Query: 479 FIITSTYQEIAGSKDRPGQYESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPY 538
FIITSTYQEIAGSKDRPGQYE+H AFT+PGLCRVVSGINVFDPKFNIAAPGADQS+YFP+
Sbjct: 543 FIITSTYQEIAGSKDRPGQYETHTAFTMPGLCRVVSGINVFDPKFNIAAPGADQSVYFPF 722
Query: 539 TEKDQRHSQFHPAIEDLLFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGK 598
TEK+QR + F PAIE LL++KV+N EHIG+L DK+KPIIFSMARLD VKN+SGLVEW+ K
Sbjct: 723 TEKNQRLTTFQPAIEGLLYSKVENEEHIGFLEDKKKPIIFSMARLDKVKNISGLVEWFAK 902
Query: 599 NKRLRNLVNLVIVGGFFDPSKSKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYR 656
NKRLR+LVNLVIVGGFFDPSKSKDREE EIKKMH L+++Y+L+GQFRWIAAQTDRYR
Sbjct: 903 NKRLRSLVNLVIVGGFFDPSKSKDREETEEIKKMHYLMKEYKLQGQFRWIAAQTDRYR 1076
Score = 35.8 bits (81), Expect(2) = e-176
Identities = 15/17 (88%), Positives = 16/17 (93%)
Frame = +2
Query: 657 NGELYRCIADTKGAFVQ 673
+GELYRC ADTKGAFVQ
Sbjct: 1076 HGELYRCXADTKGAFVQ 1126
>AW687280 similar to PIR|C96760|C96 probable sucrose synthase T9L24.42
[imported] - Arabidopsis thaliana, partial (15%)
Length = 620
Score = 118 bits (296), Expect(2) = 1e-51
Identities = 57/72 (79%), Positives = 65/72 (90%)
Frame = +2
Query: 567 GYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLRNLVNLVIVGGFFDPSKSKDREEM 626
G+L DK+KPIIFSMARLD VKN+SGLVEW+ KNKRLR+LVNLVIVGGFFDPSKSKD +E
Sbjct: 137 GFLEDKKKPIIFSMARLDKVKNISGLVEWFAKNKRLRSLVNLVIVGGFFDPSKSKDSKET 316
Query: 627 AEIKKMHDLIEK 638
EIKKMH LI++
Sbjct: 317 EEIKKMHYLIDE 352
Score = 104 bits (259), Expect(2) = 1e-51
Identities = 49/61 (80%), Positives = 54/61 (88%)
Frame = +3
Query: 635 LIEKYQLKGQFRWIAAQTDRYRNGELYRCIADTKGAFVQPALYEAFGLTVIEAMNCGLPT 694
L+ +Y+L+GQFRWIAAQTDRYRNGELYRCIADTKGAFVQPALYEAFGLTVIEA +
Sbjct: 345 LMNEYKLQGQFRWIAAQTDRYRNGELYRCIADTKGAFVQPALYEAFGLTVIEANELWITN 524
Query: 695 F 695
F
Sbjct: 525 F 527
Score = 50.4 bits (119), Expect = 3e-06
Identities = 23/31 (74%), Positives = 25/31 (80%)
Frame = +1
Query: 685 IEAMNCGLPTFATNQGGPAEIIVDGVSGFHI 715
++ MNCGLPTFATNQGGPAEIIV FHI
Sbjct: 496 LKQMNCGLPTFATNQGGPAEIIV*RCISFHI 588
>TC85404 GP|4584690|emb|CAB40794.1 sucrose synthase {Medicago truncatula},
partial (14%)
Length = 558
Score = 179 bits (455), Expect = 3e-45
Identities = 84/119 (70%), Positives = 101/119 (84%)
Frame = +2
Query: 484 TYQEIAGSKDRPGQYESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPYTEKDQ 543
T+QEIAGSKD+ GQYESH AFTLPGL RVV GI+VFDPKFNI +PGADQ+IYFPYTE +
Sbjct: 200 TFQEIAGSKDKVGQYESHTAFTLPGLYRVVHGIDVFDPKFNIVSPGADQTIYFPYTETSR 379
Query: 544 RHSQFHPAIEDLLFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRL 602
R + F+P IE+LL++ V+N EHI L D+ KPIIF+MARLD VKN++GLVEWYGKN +L
Sbjct: 380 RLTSFYPEIEELLYSSVENEEHICVLKDRNKPIIFTMARLDRVKNITGLVEWYGKNAKL 556
>TC82744 homologue to EGAD|142043|151512 second sucrose synthase {Pisum
sativum}, partial (19%)
Length = 676
Score = 120 bits (300), Expect = 3e-27
Identities = 62/153 (40%), Positives = 95/153 (61%)
Frame = +3
Query: 9 RTNSIADNMPDALRKSRYHMKKCFAKYLEKGRRIMKLHELMEEVERTIDDINERNYILEG 68
R SI D + D L R + ++Y+ +G+ I++ H L++E+E + + +++ +G
Sbjct: 225 RVPSIRDRVQDTLSAHRNELISLLSRYVAQGKGILQPHNLIDELENIL---GQEDHLKDG 395
Query: 69 NLGFILSSTQEAVVDPPYVAFAIRPNPGVWEYVRVNSEDLSVEPITPTDYLKFKERVYDQ 128
G I+ S QEA+V PP+VA A+RP PGVWEYVRVN +L+VE ++ ++YL FKE + D
Sbjct: 396 PFGEIIKSAQEAIVLPPFVAIAVRPRPGVWEYVRVNVFELNVEQLSISEYLSFKEELVDG 575
Query: 129 KWANDENAFEADFGAFDIGIPKLTLSSSIGNGL 161
K N+ E D F+ P+ T SSSIGNG+
Sbjct: 576 K-INENFVLELDLEPFNASFPRPTRSSSIGNGV 671
>BQ137165 similar to GP|4584690|emb| sucrose synthase {Medicago truncatula},
partial (9%)
Length = 968
Score = 104 bits (260), Expect = 1e-22
Identities = 50/84 (59%), Positives = 63/84 (74%)
Frame = +1
Query: 489 AGSKDRPGQYESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPYTEKDQRHSQF 548
A KD+ GQYESH AFTLPGL RV GI+VFDPKFNI +PGADQ+IYFPYTE ++ +
Sbjct: 1 ARGKDKVGQYESHTAFTLPGLYRVEHGIDVFDPKFNIESPGADQTIYFPYTETSRQLTSL 180
Query: 549 HPAIEDLLFNKVDNNEHIGYLADK 572
+P I++LL+ +N EHI L D+
Sbjct: 181 YPEIKELLYY*GENEEHIFVLKDQ 252
>TC77988 homologue to GP|18375499|gb|AAK09427.2 sucrose-phosphate synthase
{Medicago sativa}, partial (62%)
Length = 2248
Score = 101 bits (251), Expect = 1e-21
Identities = 78/249 (31%), Positives = 116/249 (46%), Gaps = 6/249 (2%)
Frame = +1
Query: 511 RVVSGINVFDPKFNIAAPGADQSIYFPYT---EKDQRHSQFHPAIEDLLFNKVDNNEHIG 567
R VS + P+ + PG + P E + HPA +D +E +
Sbjct: 70 RNVSCYGRYMPRVAVIPPGMEFHHIVPLDGDIETEPEGILDHPAPQDPPIW----SEIMR 237
Query: 568 YLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLRNLVNLVIVGGFFDPSKSKDREEMA 627
+ + RKP+I ++AR D KN++ LV+ +G+ + LR L NL ++ G D +
Sbjct: 238 FFTNPRKPVILALARPDPKKNITTLVKAFGECRPLRELANLTLIMGNRDGIDEMSSTSSS 417
Query: 628 EIKKMHDLIEKYQLKGQFRWIAAQTDRYRNG---ELYRCIADTKGAFVQPALYEAFGLTV 684
+ + LI+KY L GQ A ++ E+YR A TKG FV PA+ E FGLT+
Sbjct: 418 VLLSVLKLIDKYDLYGQ----VAYPKHHKQSDVPEIYRLAAKTKGVFVNPAIIEPFGLTL 585
Query: 685 IEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLNGDESSNKISDFFEKCKVDPSYWNVI 744
IEA GLP AT GGP +I +G +DP I+D K + W
Sbjct: 586 IEAAAYGLPMVATKNGGPVDIHRVLDNGLLVDP----HDQQSIADALLKLVSNKQLWAKC 753
Query: 745 SMAGLQRIN 753
+ GL+ I+
Sbjct: 754 RLNGLKNIH 780
>CB893717 similar to SP|O04933|SPS Sucrose-phosphate synthase 2 (EC 2.4.1.14)
(UDP-glucose-fructose- phosphate glucosyltransferase
2)., partial (22%)
Length = 843
Score = 59.3 bits (142), Expect = 5e-09
Identities = 31/69 (44%), Positives = 41/69 (58%)
Frame = +1
Query: 659 ELYRCIADTKGAFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPL 718
++YR A TKG F+ PAL E FGLT+IEA GLP AT GGP +I +G +DP
Sbjct: 16 DIYRYSAKTKGVFINPALVEPFGLTLIEAAAHGLPMVATKNGGPVDIHRALNNGLLVDPH 195
Query: 719 NGDESSNKI 727
+ +N +
Sbjct: 196 DQQAITNAL 222
>AW586383 similar to GP|3377764|gb|A nodule-enhanced sucrose synthase {Pisum
sativum}, partial (5%)
Length = 127
Score = 53.9 bits (128), Expect = 2e-07
Identities = 24/41 (58%), Positives = 28/41 (67%)
Frame = +3
Query: 435 QATIAHALEKTKYEDSDVKWKELDPKYHFSCQFMADTVAMN 475
Q TIAHALEKT Y +S + WK+ KYHF CQ AD A+N
Sbjct: 3 QCTIAHALEKTNYPESHIYWKKC*EKYHFCCQCTADLCALN 125
>BQ137986 homologue to SP|O04933|SPS Sucrose-phosphate synthase 2 (EC
2.4.1.14) (UDP-glucose-fructose- phosphate
glucosyltransferase 2)., partial (19%)
Length = 658
Score = 51.6 bits (122), Expect = 1e-06
Identities = 32/144 (22%), Positives = 74/144 (51%), Gaps = 15/144 (10%)
Frame = +3
Query: 360 DTKHSHILRVPFHTEKGILPQWVSRFDIYPYLERFTQDATTKILDLME--------GKPD 411
++ ++I+R+PF G +++ + ++P+++ F A IL++ + G+P
Sbjct: 216 ESSGAYIIRIPF----GPRDKYLEKELLWPHIQEFVDGALAHILNMSKVLGEQVGGGQPV 383
Query: 412 ---LVIGNYTDGNLVASLMARKLGITQATIAHALEKTKYED----SDVKWKELDPKYHFS 464
++ G+Y D A+L++ L + H+L + K E W++++ Y
Sbjct: 384 WPYVIHGHYADAGDSAALLSGALNVPMVLTGHSLGRNKLEQLLKQGRQSWEDINSTYKIM 563
Query: 465 CQFMADTVAMNSSDFIITSTYQEI 488
+ A+ +++++++ +ITST QEI
Sbjct: 564 RRIEAEELSLDAAELVITSTRQEI 635
>BM813415 similar to GP|20856587|gb At1g78800/F9K20_16 {Arabidopsis
thaliana}, partial (49%)
Length = 678
Score = 43.5 bits (101), Expect = 3e-04
Identities = 35/139 (25%), Positives = 61/139 (43%), Gaps = 8/139 (5%)
Frame = +1
Query: 574 KPIIFSMARLDVVKNL----SGLVEWYGKNKRLRNLV----NLVIVGGFFDPSKSKDREE 625
KP S+ R + KN+ S Y N+ L++ +L + GGF + +E
Sbjct: 283 KPNFLSINRFERKKNIQLAISAFAMLYSPNRVLKHQAITNASLTVAGGF----DKRLKEN 450
Query: 626 MAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNGELYRCIADTKGAFVQPALYEAFGLTVI 685
+ ++++ DL EK + + +++ + + RN L C+ + P FG+ +
Sbjct: 451 VEYLEELKDLAEKEGVSDKIKFVTSCSTDERNALLSECL----WCPLHPQRMSTFGIVPL 618
Query: 686 EAMNCGLPTFATNQGGPAE 704
EAM A N GGP E
Sbjct: 619 EAMAAYKVGIACNSGGPVE 675
>BE203403 similar to GP|12322785|gb unknown protein; 24439-25635 {Arabidopsis
thaliana}, partial (47%)
Length = 575
Score = 38.5 bits (88), Expect = 0.010
Identities = 17/39 (43%), Positives = 24/39 (60%)
Frame = +2
Query: 670 AFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVD 708
AFV P+ E +G ++EAM+ LP ATN GP E + +
Sbjct: 374 AFVLPSRGEGWGRPLVEAMSMSLPVIATNWSGPTEFLTE 490
>TC81003 homologue to GP|18375499|gb|AAK09427.2 sucrose-phosphate synthase
{Medicago sativa}, partial (30%)
Length = 1164
Score = 36.2 bits (82), Expect = 0.049
Identities = 33/136 (24%), Positives = 63/136 (46%), Gaps = 13/136 (9%)
Frame = +3
Query: 283 VVIFSVHGYF-GQADVLGLP-DTGGQVVYILDQVKALEEELILRIKQQGLNYKPQILVVT 340
+V+ S+HG G+ LG DTGGQV Y+++ +AL G+ Y+ +L
Sbjct: 693 IVLISIHGLIRGENMELGRDSDTGGQVKYVVELARALG-------SMPGV-YRVDLLTRQ 848
Query: 341 RLIPDA----------RGTKCHQEF-EPINDTKHSHILRVPFHTEKGILPQWVSRFDIYP 389
PD + EF + + ++ ++I+R+PF G +++ + +++P
Sbjct: 849 VASPDVDWSYGEPTEMLAPRNTDEFGDDMGESSGAYIIRIPF----GPRNKYIXKEELWP 1016
Query: 390 YLERFTQDATTKILDL 405
Y+ F A I+ +
Sbjct: 1017YIPEFVDGAIGHIIQM 1064
>TC85733 similar to GP|3832512|gb|AAC70779.1| granule-bound glycogen (starch)
synthase {Astragalus membranaceus}, partial (94%)
Length = 2251
Score = 35.8 bits (81), Expect = 0.064
Identities = 38/154 (24%), Positives = 63/154 (40%)
Frame = +1
Query: 566 IGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLRNLVNLVIVGGFFDPSKSKDREE 625
+G DK P+I + RL+ K LVE K + V ++++G + K E+
Sbjct: 1207 VGLPVDKSIPLIGFIGRLEEQKGSDILVEAIA--KFIDQNVQIIVLG-----TGKKIMEK 1365
Query: 626 MAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNGELYRCIADTKGAFVQPALYEAFGLTVI 685
E QL+ + A ++ NG L I + P+ +E GL +
Sbjct: 1366 QIE-----------QLEVTYPDKAIGVAKF-NGPLAHKIIAGADFIIIPSRFEPCGLVQL 1509
Query: 686 EAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLN 719
AM G ++ GG + + +G +GFH N
Sbjct: 1510 HAMPYGTVPIVSSTGGLVDTVKEGYTGFHAGAFN 1611
>TC84840 similar to PIR|C84671|C84671 NAM (no apical meristem)-like protein
[imported] - Arabidopsis thaliana, partial (47%)
Length = 683
Score = 32.0 bits (71), Expect = 0.93
Identities = 14/43 (32%), Positives = 25/43 (57%)
Frame = +1
Query: 718 LNGDESSNKISDFFEKCKVDPSYWNVISMAGLQRINEWYYLRP 760
L+G E S ++ E CK +P W++ + + +Q NEW++ P
Sbjct: 298 LDGHEESVQVISEVELCKYEP--WDLPAKSFIQSDNEWFFFSP 420
>BF647975 similar to PIR|T04762|T04 chitinase homolog T16H5.170 - Arabidopsis
thaliana, partial (28%)
Length = 549
Score = 31.2 bits (69), Expect = 1.6
Identities = 26/105 (24%), Positives = 49/105 (45%), Gaps = 2/105 (1%)
Frame = +2
Query: 447 YEDSDVKWKELDPKY--HFSCQFMADTVAMNSSDFIITSTYQEIAGSKDRPGQYESHAAF 504
Y DSD+ ++DP Y H C F AD + N++ I+ST+ + Q +S++
Sbjct: 170 YSDSDLAVSDIDPSYFTHLFCAF-AD-LDSNTNQVTISSTHAASFSTFTETIQSKSNSVK 343
Query: 505 TLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPYTEKDQRHSQFH 549
TL + G + N+A+ + + + + + R++ FH
Sbjct: 344 TLLSIGGGGGGPTLAQKFANMASQASTRKSFIDSSIRLARNNNFH 478
>TC89200 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (47%)
Length = 1323
Score = 30.8 bits (68), Expect = 2.1
Identities = 19/46 (41%), Positives = 27/46 (58%)
Frame = +3
Query: 195 SLMINDTLSSAAKLQMALIVADVFLSAIPKDTSYQKFELRLKEWGF 240
SL+++ T+SS Q+ +I + LS T QKF+L KEWGF
Sbjct: 246 SLVVSSTISSQ---QVPIIDLNKLLSE-EDGTELQKFDLACKEWGF 371
>BF632466
Length = 579
Score = 28.9 bits (63), Expect = 7.9
Identities = 20/54 (37%), Positives = 26/54 (48%), Gaps = 4/54 (7%)
Frame = -2
Query: 461 YHFSCQFMADTVAMNSSDFIITSTYQEIAGSKD---RPGQY-ESHAAFTLPGLC 510
YH SCQ+M A SS I+ ++ S D RPG + +S AA L C
Sbjct: 377 YHISCQYMKGLKAWESSLMILLASMSPYICS*DIWMRPGNW*QSFAAENLQNQC 216
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.138 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,581,333
Number of Sequences: 36976
Number of extensions: 295739
Number of successful extensions: 1216
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 1207
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1209
length of query: 780
length of database: 9,014,727
effective HSP length: 104
effective length of query: 676
effective length of database: 5,169,223
effective search space: 3494394748
effective search space used: 3494394748
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148342.7


