

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148291.4 + phase: 0 /pseudo
(659 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
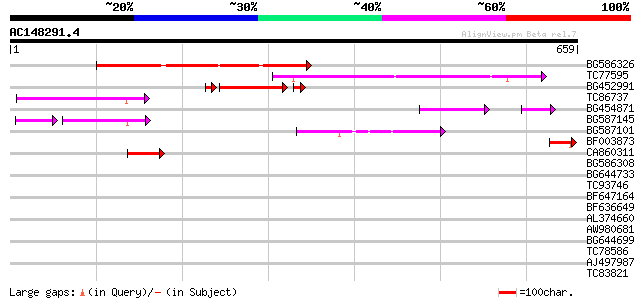
Score E
Sequences producing significant alignments: (bits) Value
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 204 9e-53
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 167 9e-42
BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, pa... 105 7e-26
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 111 1e-24
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 72 3e-16
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 64 4e-14
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 53 3e-07
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 53 3e-07
CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product ... 43 3e-04
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 37 0.024
BG644733 weakly similar to GP|15289942|db putative polyprotein {... 37 0.024
TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical... 33 0.27
BF647164 weakly similar to GP|6691191|gb F7F22.15 {Arabidopsis t... 32 1.0
BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativ... 30 2.3
AL374660 similar to GP|16518983|gb| acyl carrier protein {Olea e... 30 3.9
AW980681 similar to GP|23237931|dbj contains ESTs C26074(C11585)... 29 5.0
BG644699 similar to PIR|T07863|T078 probable polyprotein - pinea... 29 6.6
TC78586 similar to PIR|T01041|T01041 hypothetical protein YUP8H1... 29 6.6
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 29 6.6
TC83821 similar to GP|8953400|emb|CAB96673.1 1-D-deoxyxylulose 5... 28 8.6
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 204 bits (519), Expect = 9e-53
Identities = 114/251 (45%), Positives = 163/251 (64%), Gaps = 1/251 (0%)
Frame = +2
Query: 102 TIAPVLILPKPEEPFVVYCDASKLGLGGVLMQDGKVVAYASRQLRVHEKNYPTHDLELAA 161
T AP+L+LP+ +VVY DAS GLG VL Q KV+AYASRQLR HE NYPTHDLE+AA
Sbjct: 8 TSAPILVLPELIT-YVVYTDASITGLGCVLTQHEKVIAYASRQLRKHEGNYPTHDLEMAA 184
Query: 162 VVFVLKIWRHYLYGSRFEVFRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYDFGLNY 221
VVF LKIWR YLYG+ + ++ +DHKSLKY+F Q ELN+RQRRW+E + DYD + Y
Sbjct: 185 VVFALKIWRSYLYGA-----KVQIHTDHKSLKYIFTQPELNLRQRRWMEFVADYDLDITY 349
Query: 222 HPG*ANVVADALSRKTLHMSALMVKEFELLEHFRDMSLVCELSFQSIHLGMLKID-SDFL 280
+PG AN+VADALSR+ + +SA +E + L+ + L+ + LG+ ++ +D
Sbjct: 350 YPGKANLVADALSRRRVDVSA--EREADDLDGMVRALRLNVLTKATESLGLEAVNQADLF 523
Query: 281 NSIREAQKVDLKFVDLMTSGNDTEDSDFKVDDQGVLRFRGKVCIPDNEELKKLILEESHK 340
IR AQ D + + + ++++ G + G++ +P++ LK+ I+ E+HK
Sbjct: 524 TRIRLAQGQD----ENLQKVAQNDRTEYQTAKDGTILVNGRISVPNDRSLKEEIMSEAHK 691
Query: 341 SRLSIHPGATK 351
SR S+HPGA +
Sbjct: 692 SRFSVHPGAPR 724
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial
(14%)
Length = 1708
Score = 167 bits (424), Expect = 9e-42
Identities = 110/333 (33%), Positives = 168/333 (50%), Gaps = 14/333 (4%)
Frame = +2
Query: 306 SDFKVDDQGVLRFRGKVCIPDNE-------ELKKLILEESHKSRLSIHPGATKMYHDLKK 358
S+ ++D L FRG++ +P ++ EL+ +++ESH S + HPG + +
Sbjct: 74 SECQLDSLKRLTFRGRIWVPGSDDEESPLNELRTKLVQESHDSTAAGHPGRNGTLEIVSR 253
Query: 359 LFWWSGLKRDVAQFVYACLICQKSKVEHQKPAGLLTPLDVPEWKWDSISMDFVTSLPNTP 418
F+W G + V +FV C +C + Q G L PL VP +SMDF+TSLP T
Sbjct: 254 KFFWPGQSQTVRRFVRNCDVCGGIHIWRQAKRGFLKPLPVPNRLHSDLSMDFITSLPPTR 433
Query: 419 -RGHDAIWVVVDRLTKSAHFIPININYPVAQLAEIYIHSVVKLHGVPASIVSDRDPRFTS 477
RG +WV+VDRL+KS ++ A A+ ++ + HG+P SIVSDR +
Sbjct: 434 GRGSQYLWVIVDRLSKSVTLEEMDTMEAEA-CAQRFLSCHYRFHGMPQSIVSDRGSNWVG 610
Query: 478 RFWKRLQDALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGAWDSHLPLIEFT 537
RFW+ G LS++YHPQTDG +ER Q ++ +LR V W LP ++
Sbjct: 611 RFWREFCRLTGVTQLLSTSYHPQTDGGTERWNQEIQAVLRAYVCWSQDNWGDLLPTVQLA 790
Query: 538 YNNSYHSSIGMAPFEALYGRRCRTPLCWFESGESVVLGPD-----LVHETTEKVRLIRER 592
N ++SSIG PF +G P+ E VV + LV + I+
Sbjct: 791 LRNRHNSSIGATPFFVEHGYHV-DPIPTVEDTGGVVSEGEAAAQLLVKRMKDVTGFIQAE 967
Query: 593 MKASQSRQKSYHDKRRKELE-FQEGDHVFLRVT 624
+ A+Q R ++ +KRR + +Q GD V+L V+
Sbjct: 968 IVAAQQRSEASANKRRCPADRYQVGDKVWLNVS 1066
>BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, partial (33%)
Length = 560
Score = 105 bits (263), Expect(3) = 7e-26
Identities = 50/78 (64%), Positives = 65/78 (83%)
Frame = +3
Query: 245 VKEFELLEHFRDMSLVCELSFQSIHLGMLKIDSDFLNSIREAQKVDLKFVDLMTSGNDTE 304
++ + LE FRD+SLVCE+S QS+ LGMLKI+++FL+SI+EAQKVD+K VDLM N TE
Sbjct: 51 LESWSCLEQFRDLSLVCEVSPQSVKLGMLKINNEFLDSIKEAQKVDVKLVDLMFGNNQTE 230
Query: 305 DSDFKVDDQGVLRFRGKV 322
D DFKVDDQGVL+FR ++
Sbjct: 231 DGDFKVDDQGVLQFRDRI 284
Score = 25.4 bits (54), Expect(3) = 7e-26
Identities = 11/13 (84%), Positives = 12/13 (91%)
Frame = +1
Query: 228 VVADALSRKTLHM 240
VVAD LSRKTLH+
Sbjct: 1 VVADVLSRKTLHV 39
Score = 24.6 bits (52), Expect(3) = 7e-26
Identities = 9/15 (60%), Positives = 14/15 (93%)
Frame = +2
Query: 330 LKKLILEESHKSRLS 344
+KK+ILEESH+S ++
Sbjct: 284 MKKMILEESHRSNVN 328
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 111 bits (277), Expect = 1e-24
Identities = 67/161 (41%), Positives = 89/161 (54%), Gaps = 6/161 (3%)
Frame = +1
Query: 8 KCEFWLSEVSFLGHIIF-GSDIAVDPSKVDAVSQWETLKSVTEIRSFLGLAGYYRRFIEG 66
KCEF ++ V ++G I+ G ++ DP K+ A+ W SV RSFLG YY+ FI G
Sbjct: 1030 KCEFSVTTVKYVGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGARSFLGFCNYYKDFIPG 1209
Query: 67 FSKLALPSTQLTCKGKSFVWDAQCERSFNELKRRLTIAPVLILPKPEEPFVVYCDASKLG 126
+S++ P T+LT K F W A+ E +F +LKR PVL + PE V D S
Sbjct: 1210 YSEITEPLTRLTRKDFPFRWGAEQEAAFTKLKRLFAEEPVLRMFDPEAVTTVETDCSGFA 1389
Query: 127 LGGVLMQD-----GKVVAYASRQLRVHEKNYPTHDLELAAV 162
LGGVL Q+ VA+ S++L E NYP HD EL AV
Sbjct: 1390 LGGVLTQEDGTGAAHPVAFHSQRLSPAEYNYPIHDKELLAV 1512
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 72.0 bits (175), Expect(2) = 3e-16
Identities = 37/81 (45%), Positives = 46/81 (56%)
Frame = +2
Query: 477 SRFWKRLQDALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGAWDSHLPLIEF 536
S FWK+L G+ L +SSAYHP +DGQSE + E LR + W P E+
Sbjct: 32 SNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEY 211
Query: 537 TYNNSYHSSIGMAPFEALYGR 557
YN SY+ S M PF+ALYGR
Sbjct: 212 WYNTSYNISAAMTPFKALYGR 274
Score = 31.6 bits (70), Expect(2) = 3e-16
Identities = 14/39 (35%), Positives = 22/39 (55%)
Frame = +1
Query: 596 SQSRQKSYHDKRRKELEFQEGDHVFLRVTPLTGVGRALK 634
+Q K DK+R+ EFQ G+HV +++ P AL+
Sbjct: 388 AQQTMKHQADKKRRHFEFQLGEHVLVKLQPYQQSSVALR 504
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 64.3 bits (155), Expect(2) = 4e-14
Identities = 36/106 (33%), Positives = 55/106 (50%), Gaps = 4/106 (3%)
Frame = +3
Query: 62 RFIEGFSKLALPSTQLTCKGKSFVWDAQCERSFNELKRRLTIAPVLILPKPEEPFVVYCD 121
RFI + LP +L C K FVWD +CE +F +LK+ LT PVL P+ + +Y
Sbjct: 429 RFISRSTDKCLPFYKLLCGNKRFVWDEKCEEAFEQLKQYLTTPPVLSKPEAGDTLSLYIA 608
Query: 122 ASKLGLGGVLMQDG----KVVAYASRQLRVHEKNYPTHDLELAAVV 163
S + VL+++ K + Y S+++ E YPT + AV+
Sbjct: 609 ISSTAVSSVLIREDRGEQKPIFYTSKRMTDPETRYPTLEKMAFAVI 746
Score = 32.0 bits (71), Expect(2) = 4e-14
Identities = 14/49 (28%), Positives = 27/49 (54%)
Frame = +2
Query: 7 SKCEFWLSEVSFLGHIIFGSDIAVDPSKVDAVSQWETLKSVTEIRSFLG 55
+KC F ++ FLG+I+ I V+P ++ A+ + K+ E++ G
Sbjct: 263 AKCTFGVTSGEFLGYIVTQQGIEVNPKQITAILDLPSPKNSREVQRLTG 409
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 53.1 bits (126), Expect = 3e-07
Identities = 41/177 (23%), Positives = 82/177 (46%), Gaps = 4/177 (2%)
Frame = +2
Query: 334 ILEESHKSRLSIHPGATKMYHDLKKL-FWWSGLKRDVAQFVYACLICQKS---KVEHQKP 389
IL H S + H +K +++ FWW + +D F+ C CQ+ ++ P
Sbjct: 104 ILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHSFISKCDPCQRQGNIS*RNEMP 283
Query: 390 AGLLTPLDVPEWKWDSISMDFVTSLPNTPRGHDAIWVVVDRLTKSAHFIPININYPVAQL 449
+ ++V +D +DF+ P++ + I V VD ++K I N +
Sbjct: 284 QNFILEVEV----FDVWGIDFMGPFPSS-YNNKYILVAVDYVSKWVEAIASPTN-DATVV 445
Query: 450 AEIYIHSVVKLHGVPASIVSDRDPRFTSRFWKRLQDALGSKLRLSSAYHPQTDGQSE 506
+++ + GVP ++SD F ++ +++L G + ++++AYHPQ +S+
Sbjct: 446 VKMFKSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHKVATAYHPQKAERSK 616
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 53.1 bits (126), Expect = 3e-07
Identities = 27/38 (71%), Positives = 30/38 (78%), Gaps = 7/38 (18%)
Frame = +2
Query: 628 GVGRALKSRKLTPKFIGPYQISD-------RVGLPPHL 658
GVGRALKS+KLT +FIGPYQIS+ RVGLPPHL
Sbjct: 2 GVGRALKSKKLTVRFIGPYQISERVGTVAYRVGLPPHL 115
>CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product {Drosophila
melanogaster}, partial (20%)
Length = 192
Score = 43.1 bits (100), Expect = 3e-04
Identities = 18/43 (41%), Positives = 29/43 (66%)
Frame = +1
Query: 138 VAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEV 180
+AYASR L E+NY + E A ++ ++ +RHYL+G +FE+
Sbjct: 64 IAYASRLLTAAERNYTVVERECLAAIWAIRNFRHYLHGPKFEL 192
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 686
Score = 37.0 bits (84), Expect = 0.024
Identities = 20/71 (28%), Positives = 37/71 (51%)
Frame = -2
Query: 461 HGVPASIVSDRDPRFTSRFWKRLQDALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCV 520
HG+P IV+D F S ++ + +L +S +PQ++GQ+E + + + D L+ +
Sbjct: 685 HGLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKIIIDGLKKRL 506
Query: 521 LEQGGAWDSHL 531
+ G W L
Sbjct: 505 DLKKGCWADEL 473
>BG644733 weakly similar to GP|15289942|db putative polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (1%)
Length = 174
Score = 37.0 bits (84), Expect = 0.024
Identities = 18/43 (41%), Positives = 26/43 (59%)
Frame = -3
Query: 419 RGHDAIWVVVDRLTKSAHFIPININYPVAQLAEIYIHSVVKLH 461
+ +++I VVVDRLTKS FIP +Y A I + +V +H
Sbjct: 130 KSYESI*VVVDRLTKSTLFIPFKTSYSAK*YARILLDEIVCIH 2
>TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical
protein~similar to gag-pol polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (4%)
Length = 1019
Score = 33.5 bits (75), Expect = 0.27
Identities = 24/84 (28%), Positives = 39/84 (45%), Gaps = 13/84 (15%)
Frame = +2
Query: 368 DVAQFVYACLICQKSKVEHQKPAGLLTPLD-------------VPEWKWDSISMDFVTSL 414
D + +A L + K E KP G L ++ VP+ W+ +++DF L
Sbjct: 350 DAIKIKFAPLPLNEGKEEF-KPLGFLADMEPFKDKTKLCLLRPVPKPPWEDVTIDFSLGL 526
Query: 415 PNTPRGHDAIWVVVDRLTKSAHFI 438
T + D+ VV D+ ++ AHFI
Sbjct: 527 L*TQQLKDSKMVVGDKFSRMAHFI 598
>BF647164 weakly similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana},
partial (6%)
Length = 469
Score = 31.6 bits (70), Expect = 1.0
Identities = 23/68 (33%), Positives = 35/68 (50%), Gaps = 1/68 (1%)
Frame = +1
Query: 584 EKVRL-IRERMKASQSRQKSYHDKRRKELEFQEGDHVFLRVTPLTGVGRALKSRKLTPKF 642
E++RL E K + R K +HD+R EF+EG+ V L + L L KL +
Sbjct: 118 EELRLDAYENAKIYKERTKKWHDRRIIRREFREGELVLLFNSRL-----KLFPGKLRSHW 282
Query: 643 IGPYQISD 650
GP+Q+ +
Sbjct: 283 SGPFQVKN 306
>BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativa}, partial
(4%)
Length = 653
Score = 30.4 bits (67), Expect = 2.3
Identities = 17/55 (30%), Positives = 26/55 (46%)
Frame = +1
Query: 501 TDGQSERTIQSLEDLLRVCVLEQGGAWDSHLPLIEFTYNNSYHSSIGMAPFEALY 555
+D Q+ +LE L EQ G + L E YN ++H + G PF+ +Y
Sbjct: 7 SDEQAGLLNHTLETHLLYFTSEQQGV*NFFLTWAECLYNTNFHRTAGCTPFKVVY 171
>AL374660 similar to GP|16518983|gb| acyl carrier protein {Olea europaea},
partial (45%)
Length = 363
Score = 29.6 bits (65), Expect = 3.9
Identities = 10/17 (58%), Positives = 14/17 (81%)
Frame = +1
Query: 168 IWRHYLYGSRFEVFRFE 184
IW+ +L+G RF VF+FE
Sbjct: 262 IWKFFLFGKRFSVFQFE 312
>AW980681 similar to GP|23237931|dbj contains ESTs C26074(C11585)
AU092275(C11585)~unknown protein, partial (11%)
Length = 748
Score = 29.3 bits (64), Expect = 5.0
Identities = 12/25 (48%), Positives = 16/25 (64%)
Frame = +2
Query: 6 LSKCEFWLSEVSFLGHIIFGSDIAV 30
LSK FW S + FL H+ F +D+ V
Sbjct: 254 LSKVSFWASSIHFLYHVQFHNDLHV 328
>BG644699 similar to PIR|T07863|T078 probable polyprotein - pineapple
retrotransposon dea1 (fragment), partial (5%)
Length = 231
Score = 28.9 bits (63), Expect = 6.6
Identities = 19/51 (37%), Positives = 28/51 (54%), Gaps = 8/51 (15%)
Frame = +2
Query: 617 DHVFLRVTPLT-GVGRALKSRKLTPKFIGPYQISDRVG-------LPPHLS 659
+ V L+V P G R K KL+ ++IGP+++ R+G LPP LS
Sbjct: 2 EQVLLKVLPTERGDCRFGKRGKLSLRYIGPFEVIKRIGEVAYELALPPGLS 154
>TC78586 similar to PIR|T01041|T01041 hypothetical protein YUP8H12R.24 -
Arabidopsis thaliana, partial (38%)
Length = 2831
Score = 28.9 bits (63), Expect = 6.6
Identities = 25/93 (26%), Positives = 42/93 (44%)
Frame = +1
Query: 220 NYHPG*ANVVADALSRKTLHMSALMVKEFELLEHFRDMSLVCELSFQSIHLGMLKIDSDF 279
+YHP + +A LS + + + + + L F+DMS+ ELSF+ G +K+++
Sbjct: 2176 HYHPA-ISTMATGLSSMSTEQNQVFLSKSSPLLAFKDMSIDQELSFE--QSGSIKLNNKR 2346
Query: 280 LNSIREAQKVDLKFVDLMTSGNDTEDSDFKVDD 312
S A + G+ T S F DD
Sbjct: 2347 KRSHGNATSDSI--------GSTTVTSSFNEDD 2421
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 28.9 bits (63), Expect = 6.6
Identities = 23/81 (28%), Positives = 38/81 (46%), Gaps = 3/81 (3%)
Frame = -2
Query: 161 AVVFVLKIWRHYLYGSRFEVFRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYDFGLN 220
A+ + K RHY+ + S +KY+F++ L R RW LL +YD +
Sbjct: 623 ALAWAAKRLRHYMINHTTWLV-----SKMDPIKYIFEKPALTGRIARWQMLLSEYD--IE 465
Query: 221 YHPG*A---NVVADALSRKTL 238
Y A +++AD L+ + L
Sbjct: 464 YRSQKAIKGSILADHLAHQPL 402
>TC83821 similar to GP|8953400|emb|CAB96673.1 1-D-deoxyxylulose 5-phosphate
synthase-like protein {Arabidopsis thaliana}, partial
(46%)
Length = 1172
Score = 28.5 bits (62), Expect = 8.6
Identities = 14/36 (38%), Positives = 20/36 (54%)
Frame = -3
Query: 403 WDSISMDFVTSLPNTPRGHDAIWVVVDRLTKSAHFI 438
W+SI FV TPR A W+V+D ++ S H +
Sbjct: 762 WNSIKYSFVFPNQGTPRIPKANWLVID-MSSSRHHV 658
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.139 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,247,623
Number of Sequences: 36976
Number of extensions: 297843
Number of successful extensions: 1368
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 1358
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1361
length of query: 659
length of database: 9,014,727
effective HSP length: 102
effective length of query: 557
effective length of database: 5,243,175
effective search space: 2920448475
effective search space used: 2920448475
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148291.4


