

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148289.7 - phase: 0
(197 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
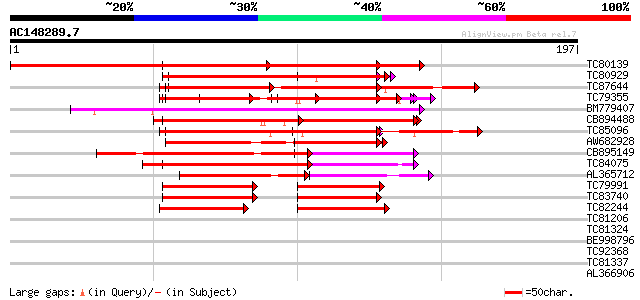
Score E
Sequences producing significant alignments: (bits) Value
TC80139 homologue to PIR|T48868|T48868 zinc finger protein [impo... 310 2e-85
TC80929 similar to GP|10177307|dbj|BAB10568. contains similarity... 126 5e-30
TC87644 similar to GP|10177307|dbj|BAB10568. contains similarity... 124 2e-29
TC79355 similar to GP|15081721|gb|AAK82515.1 At1g04990/F13M7_1 {... 114 2e-26
BM779407 similar to GP|6437560|gb|A hypothetical protein {Arabid... 107 2e-24
CB894488 similar to GP|6437560|gb| hypothetical protein {Arabido... 94 3e-20
TC85096 similar to GP|22507458|gb|AAH19429.1 Unknown (protein fo... 91 2e-19
AW682928 similar to GP|22507458|gb| Unknown (protein for MGC:303... 89 1e-18
CB895149 similar to GP|6437560|gb|A hypothetical protein {Arabid... 87 5e-18
TC84075 similar to GP|3738297|gb|AAC63639.1| unknown protein {Ar... 84 5e-17
AL365712 similar to GP|3738297|gb|A unknown protein {Arabidopsis... 69 9e-13
TC79991 similar to GP|10177307|dbj|BAB10568. contains similarity... 52 1e-07
TC83740 similar to GP|6437560|gb|AAF08587.1| hypothetical protei... 52 1e-07
TC82244 similar to GP|22507458|gb|AAH19429.1 Unknown (protein fo... 44 3e-05
TC81206 similar to GP|8777433|dbj|BAA97023.1 gene_id:MHM17.4~unk... 39 0.001
TC81324 similar to GP|21553871|gb|AAM62964.1 unknown {Arabidopsi... 37 0.006
BE998796 similar to GP|5441893|dbj| ESTs C99174(E10437) D22295(C... 33 0.055
TC92368 weakly similar to GP|19347974|gb|AAL86319.1 unknown prot... 33 0.072
TC81337 similar to GP|15723293|gb|AAL06332.1 U2 auxiliary factor... 32 0.16
AL366906 weakly similar to PIR|T45743|T45 hypothetical protein F... 31 0.27
>TC80139 homologue to PIR|T48868|T48868 zinc finger protein [imported] -
garden pea, partial (87%)
Length = 1402
Score = 310 bits (794), Expect = 2e-85
Identities = 142/144 (98%), Positives = 143/144 (98%)
Frame = +3
Query: 1 MGSDSPQQTMRNDQTYGTSHQGDPENAGLQGVYSQYRSGSVPVGFYALQRENIFPERPDQ 60
MGSDSPQQTMRNDQTYGTSHQGDPENAGLQGVYSQYRSGSVPVGFYALQRENIFPERPDQ
Sbjct: 969 MGSDSPQQTMRNDQTYGTSHQGDPENAGLQGVYSQYRSGSVPVGFYALQRENIFPERPDQ 1148
Query: 61 PECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSRYGICKF 120
PECQFYMKTGDCKFGAVCRFHHPRERTIPAPDC LSPLGLPLRPGEPLCVFYSR+GICKF
Sbjct: 1149 PECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCDLSPLGLPLRPGEPLCVFYSRHGICKF 1328
Query: 121 GPSCKFDHPMGIFTYNVSASPLAE 144
GPSCKFDHPMGIFTYNVSASPLAE
Sbjct: 1329 GPSCKFDHPMGIFTYNVSASPLAE 1400
Score = 105 bits (261), Expect = 1e-23
Identities = 45/76 (59%), Positives = 53/76 (69%)
Frame = +3
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYS 113
FPER QPEC +Y+KTG CKFGA CRFHHP+++ A L+ LG PLRP E C +Y
Sbjct: 519 FPERLGQPECHYYLKTGTCKFGATCRFHHPKDKAGVAGRVALNILGYPLRPNESECAYYL 698
Query: 114 RYGICKFGPSCKFDHP 129
R G CKFG +CKF HP
Sbjct: 699 RTGQCKFGNTCKFHHP 746
Score = 82.0 bits (201), Expect = 1e-16
Identities = 33/76 (43%), Positives = 48/76 (62%)
Frame = +3
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYS 113
+PE P +P+C +Y++TG C+FGA CRF+HP R + + P R G+P C +Y
Sbjct: 384 YPEHPGEPDCSYYIRTGLCRFGATCRFNHPPNRKLAIATARMKG-EFPERLGQPECHYYL 560
Query: 114 RYGICKFGPSCKFDHP 129
+ G CKFG +C+F HP
Sbjct: 561 KTGTCKFGATCRFHHP 608
Score = 53.5 bits (127), Expect = 5e-08
Identities = 18/38 (47%), Positives = 27/38 (70%)
Frame = +3
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAP 91
+P RP++ EC +Y++TG CKFG C+FHHP+ + P
Sbjct: 657 YPLRPNESECAYYLRTGQCKFGNTCKFHHPQPSNMVLP 770
>TC80929 similar to GP|10177307|dbj|BAB10568. contains similarity to zinc
finger protein~gene_id:MDC12.23 {Arabidopsis thaliana},
partial (28%)
Length = 1274
Score = 126 bits (317), Expect = 5e-30
Identities = 50/79 (63%), Positives = 62/79 (78%)
Frame = +3
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYS 113
FPERP +PEC F++KTGDCKF + C+FHHP+ R P C LS GLPLRPG+ +C YS
Sbjct: 603 FPERPGEPECSFFLKTGDCKFKSHCKFHHPKNRITKLPPCNLSDKGLPLRPGQNVCTHYS 782
Query: 114 RYGICKFGPSCKFDHPMGI 132
RYGICKFGP+CK+DHP+ +
Sbjct: 783 RYGICKFGPACKYDHPINL 839
Score = 60.5 bits (145), Expect = 4e-10
Identities = 30/76 (39%), Positives = 41/76 (53%), Gaps = 2/76 (2%)
Frame = +2
Query: 56 ERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPG--EPLCVFYS 113
ER Q EC++Y ++G CKFG C+F H R + + + L P E + V
Sbjct: 47 ERSSQTECKYYSRSGGCKFGKDCKFDHTRRKIFSEIEVFRA*LPWPTYSFG*ERMPVITM 226
Query: 114 RYGICKFGPSCKFDHP 129
R G CKFG +CKF+HP
Sbjct: 227 RTGSCKFGANCKFNHP 274
Score = 42.7 bits (99), Expect = 9e-05
Identities = 18/34 (52%), Positives = 20/34 (57%)
Frame = +3
Query: 101 PLRPGEPLCVFYSRYGICKFGPSCKFDHPMGIFT 134
P RPGEP C F+ + G CKF CKF HP T
Sbjct: 606 PERPGEPECSFFLKTGDCKFKSHCKFHHPKNRIT 707
Score = 35.0 bits (79), Expect = 0.019
Identities = 13/26 (50%), Positives = 18/26 (69%)
Frame = +2
Query: 67 MKTGDCKFGAVCRFHHPRERTIPAPD 92
M+TG CKFGA C+F+HP ++ D
Sbjct: 224 MRTGSCKFGANCKFNHPDPTSVGGYD 301
>TC87644 similar to GP|10177307|dbj|BAB10568. contains similarity to zinc
finger protein~gene_id:MDC12.23 {Arabidopsis thaliana},
partial (36%)
Length = 1541
Score = 124 bits (312), Expect = 2e-29
Identities = 58/119 (48%), Positives = 76/119 (63%), Gaps = 8/119 (6%)
Frame = +1
Query: 53 IFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFY 112
+FPERP +PEC F++KTGDCKF + C+FHHP+ R P C LS GLPLRP + +C Y
Sbjct: 928 VFPERPGEPECSFFIKTGDCKFKSNCKFHHPKNRVAKLPPCNLSDKGLPLRPDQSVCSHY 1107
Query: 113 SRYGICKFGPSCKFDHP--------MGIFTYNVSASPLAEAAGRRLLGSSSGTAALSLS 163
SRYGICKFGP+C+FDHP G+ + + S A+ AG +G S+ A + S
Sbjct: 1108 SRYGICKFGPACRFDHPESALPLMMPGLGQQSFANSANAQVAG---MGGSASDATIQQS 1275
Score = 81.3 bits (199), Expect = 2e-16
Identities = 35/74 (47%), Positives = 49/74 (65%)
Frame = +1
Query: 56 ERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSRY 115
E Q EC++Y ++G CKFG C+++H R T P + L+ LGLP+R GE C +Y R
Sbjct: 403 ENAGQTECKYYQRSGGCKFGKACKYNHSRGFTAPISE--LNFLGLPIRLGERECPYYMRT 576
Query: 116 GICKFGPSCKFDHP 129
G CKFG +C+F+HP
Sbjct: 577 GSCKFGSNCRFNHP 618
Score = 48.1 bits (113), Expect = 2e-06
Identities = 19/38 (50%), Positives = 25/38 (65%)
Frame = +1
Query: 55 PERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPD 92
P R + EC +YM+TG CKFG+ CRF+HP T+ D
Sbjct: 532 PIRLGERECPYYMRTGSCKFGSNCRFNHPDPTTVGGSD 645
Score = 37.0 bits (84), Expect = 0.005
Identities = 16/34 (47%), Positives = 22/34 (64%)
Frame = +1
Query: 105 GEPLCVFYSRYGICKFGPSCKFDHPMGIFTYNVS 138
G+ C +Y R G CKFG +CK++H G FT +S
Sbjct: 412 GQTECKYYQRSGGCKFGKACKYNHSRG-FTAPIS 510
>TC79355 similar to GP|15081721|gb|AAK82515.1 At1g04990/F13M7_1 {Arabidopsis
thaliana}, partial (42%)
Length = 1627
Score = 114 bits (285), Expect = 2e-26
Identities = 44/82 (53%), Positives = 63/82 (76%)
Frame = +2
Query: 55 PERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSR 114
P+RP+QP+C+++M TG CK+G+ C+FHHP+ER A ++PLGLP+RPG +C +Y
Sbjct: 998 PDRPEQPDCKYFMSTGTCKYGSDCKFHHPKERI--AQTLSINPLGLPMRPGNAICSYYRI 1171
Query: 115 YGICKFGPSCKFDHPMGIFTYN 136
YG+CKFGP+CKFDHP+ + N
Sbjct: 1172 YGVCKFGPTCKFDHPVVAISQN 1237
Score = 72.8 bits (177), Expect = 8e-14
Identities = 30/81 (37%), Positives = 50/81 (61%), Gaps = 5/81 (6%)
Frame = +1
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPL-----GLPLRPGEPL 108
+P+RP +P+C +Y++TG C +G+ CR++HP +SP+ LP R G+P
Sbjct: 316 YPDRPGEPDCVYYLRTGMCGYGSNCRYNHPAN---------ISPVTQYGEELPERVGQPD 468
Query: 109 CVFYSRYGICKFGPSCKFDHP 129
C ++ + G CK+G +CK+ HP
Sbjct: 469 CEYFLKTGTCKYGSTCKYHHP 531
Score = 67.0 bits (162), Expect = 4e-12
Identities = 26/54 (48%), Positives = 40/54 (73%)
Frame = +1
Query: 55 PERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPL 108
PER QP+C++++KTG CK+G+ C++HHP++R AP V + LGLP+R G +
Sbjct: 445 PERVGQPDCEYFLKTGTCKYGSTCKYHHPKDRRGAAP-VVFNTLGLPMRQGRKI 603
Score = 50.4 bits (119), Expect = 4e-07
Identities = 22/57 (38%), Positives = 31/57 (53%)
Frame = +1
Query: 92 DCVLSPLGLPLRPGEPLCVFYSRYGICKFGPSCKFDHPMGIFTYNVSASPLAEAAGR 148
D V + P RPGEP CV+Y R G+C +G +C+++HP I L E G+
Sbjct: 292 DAVPQSMPYPDRPGEPDCVYYLRTGMCGYGSNCRYNHPANISPVTQYGEELPERVGQ 462
Score = 49.3 bits (116), Expect = 1e-06
Identities = 16/33 (48%), Positives = 24/33 (72%)
Frame = +2
Query: 53 IFPERPDQPECQFYMKTGDCKFGAVCRFHHPRE 85
+FP ++ C +YM+TG CKFG C+FHHP++
Sbjct: 575 VFPCVKEEKSCPYYMRTGSCKFGVACKFHHPQQ 673
Score = 45.4 bits (106), Expect = 1e-05
Identities = 25/76 (32%), Positives = 35/76 (45%)
Frame = +2
Query: 67 MKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSRYGICKFGPSCKF 126
M TG V + +P T+ + S LP RP +P C ++ G CK+G CKF
Sbjct: 902 MPTGFIGSNLVYDYMNPAGETLSGGQAMNS--SLPDRPEQPDCKYFMSTGTCKYGSDCKF 1075
Query: 127 DHPMGIFTYNVSASPL 142
HP +S +PL
Sbjct: 1076HHPKERIAQTLSINPL 1123
Score = 40.0 bits (92), Expect = 6e-04
Identities = 24/57 (42%), Positives = 28/57 (49%), Gaps = 9/57 (15%)
Frame = +2
Query: 94 VLSPLG-----LPLRPGEPLCVFYSRYGICKFGPSCKFDHPMGIFT----YNVSASP 141
VL PL P E C +Y R G CKFG +CKF HP + Y V+ASP
Sbjct: 545 VLHPLSSIL*VFPCVKEEKSCPYYMRTGSCKFGVACKFHHPQQAASFGGAYPVAASP 715
>BM779407 similar to GP|6437560|gb|A hypothetical protein {Arabidopsis
thaliana}, partial (16%)
Length = 532
Score = 107 bits (268), Expect = 2e-24
Identities = 57/137 (41%), Positives = 73/137 (52%), Gaps = 14/137 (10%)
Frame = +3
Query: 22 GDPENAG---LQGVYSQYRSGSVPVGFYAL-----------QRENIFPERPDQPECQFYM 67
G P N G L G+ G+ G Y L Q E FP P+QPE +Y
Sbjct: 96 GSPANVGSTQLYGITQLPSPGNAYTGPYQLSGSSVGPSSRNQNEQSFPASPNQPEYHYYS 275
Query: 68 KTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSRYGICKFGPSCKFD 127
K + F R+H P + + P + VLSP GLPLRPG LC Y++ GICKFGP+CKFD
Sbjct: 276 KPEELPFAPSYRYHKPPDMSAPKVNAVLSPAGLPLRPGAALCTHYAQRGICKFGPACKFD 455
Query: 128 HPMGIFTYNVSASPLAE 144
HP+ +Y+ SAS L +
Sbjct: 456 HPIAPLSYSPSASSLTD 506
>CB894488 similar to GP|6437560|gb| hypothetical protein {Arabidopsis
thaliana}, partial (37%)
Length = 830
Score = 94.4 bits (233), Expect = 3e-20
Identities = 42/90 (46%), Positives = 57/90 (62%), Gaps = 1/90 (1%)
Frame = +1
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRER-TIPAPDCVLSPLGLPLRPGEPLCVFY 112
+PER QP CQ+YM+T CKFGA C++HHP++ A L+ G PLRPGE C ++
Sbjct: 394 YPERVGQPVCQYYMRTRSCKFGASCKYHHPKQTGATDASPVSLNYYGYPLRPGEKECSYF 573
Query: 113 SRYGICKFGPSCKFDHPMGIFTYNVSASPL 142
+ G CKFG +CKFDHP+ + SP+
Sbjct: 574 VKTGQCKFGATCKFDHPVPASVQIPAPSPV 663
Score = 85.5 bits (210), Expect = 1e-17
Identities = 39/93 (41%), Positives = 57/93 (60%)
Frame = +1
Query: 51 ENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCV 110
E+ +P+RPD+ +C +Y++TG C FG+ CRF+HPR+R + P R G+P+C
Sbjct: 247 EDSYPQRPDEVDCTYYLRTGFCGFGSRCRFNHPRDRAAVIGAASRTVGEYPERVGQPVCQ 426
Query: 111 FYSRYGICKFGPSCKFDHPMGIFTYNVSASPLA 143
+Y R CKFG SCK+ HP T ASP++
Sbjct: 427 YYMRTRSCKFGASCKYHHPKQ--TGATDASPVS 519
Score = 52.8 bits (125), Expect = 9e-08
Identities = 25/53 (47%), Positives = 35/53 (65%), Gaps = 4/53 (7%)
Frame = +1
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERT--IPAPDCV--LSPLGLPL 102
+P RP + EC +++KTG CKFGA C+F HP + IPAP V +S L +P+
Sbjct: 535 YPLRPGEKECSYFVKTGQCKFGATCKFDHPVPASVQIPAPSPVPPVSSLHVPV 693
>TC85096 similar to GP|22507458|gb|AAH19429.1 Unknown (protein for
MGC:30371) {Mus musculus}, partial (21%)
Length = 834
Score = 91.3 bits (225), Expect = 2e-19
Identities = 40/81 (49%), Positives = 52/81 (63%), Gaps = 4/81 (4%)
Frame = +1
Query: 53 IFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIP----APDCVLSPLGLPLRPGEPL 108
I+P+RP Q EC FYMK+G CKFG C++HHP +RT + L+ GLP R G +
Sbjct: 265 IYPQRPGQIECDFYMKSGICKFGERCKYHHPIDRTTSLSKLQSNVKLTAAGLPRREGVEI 444
Query: 109 CVFYSRYGICKFGPSCKFDHP 129
C +Y + CKFG +CKFDHP
Sbjct: 445 CPYYLKTATCKFGATCKFDHP 507
Score = 74.7 bits (182), Expect = 2e-14
Identities = 41/116 (35%), Positives = 49/116 (41%), Gaps = 40/116 (34%)
Frame = +1
Query: 55 PERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGL-------------- 100
P RP + +C FYMKTG CKFGA CR++HP I P L P L
Sbjct: 13 PVRPREVDCPFYMKTGSCKFGASCRYNHPDMNAINPPVSALGPSVLASSTANLNMGAINP 192
Query: 101 --------------------------PLRPGEPLCVFYSRYGICKFGPSCKFDHPM 130
P RPG+ C FY + GICKFG CK+ HP+
Sbjct: 193 AASFYQAFDPSLSNPMSQVGVTGTIYPQRPGQIECDFYMKSGICKFGERCKYHHPI 360
Score = 48.1 bits (113), Expect = 2e-06
Identities = 28/67 (41%), Positives = 41/67 (60%), Gaps = 1/67 (1%)
Frame = +1
Query: 99 GLPLRPGEPLCVFYSRYGICKFGPSCKFDHPMGIFTYNVSA-SPLAEAAGRRLLGSSSGT 157
GLP+RP E C FY + G CKFG SC+++HP +++A +P A G +L SS T
Sbjct: 7 GLPVRPREVDCPFYMKTGSCKFGASCRYNHP------DMNAINPPVSALGPSVLASS--T 162
Query: 158 AALSLSS 164
A L++ +
Sbjct: 163 ANLNMGA 183
>AW682928 similar to GP|22507458|gb| Unknown (protein for MGC:30371) {Mus
musculus}, partial (13%)
Length = 641
Score = 89.0 bits (219), Expect = 1e-18
Identities = 39/77 (50%), Positives = 52/77 (66%)
Frame = +3
Query: 55 PERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSR 114
PERP +P+C F++KT CKFG C+F+HP +P+ + +S GLP RP EP C FY +
Sbjct: 363 PERPGEPDCPFFLKTQKCKFGTKCKFNHPN---VPSENDDVS--GLPERPQEPPCAFYLK 527
Query: 115 YGICKFGPSCKFDHPMG 131
G CK+G +CKF HP G
Sbjct: 528 TGKCKYGAACKFHHPQG 578
Score = 44.7 bits (104), Expect = 2e-05
Identities = 18/30 (60%), Positives = 21/30 (70%)
Frame = +3
Query: 100 LPLRPGEPLCVFYSRYGICKFGPSCKFDHP 129
LP RPGEP C F+ + CKFG CKF+HP
Sbjct: 360 LPERPGEPDCPFFLKTQKCKFGTKCKFNHP 449
>CB895149 similar to GP|6437560|gb|A hypothetical protein {Arabidopsis
thaliana}, partial (12%)
Length = 860
Score = 86.7 bits (213), Expect = 5e-18
Identities = 42/75 (56%), Positives = 51/75 (68%)
Frame = +2
Query: 31 GVYSQYRSGSVPVGFYALQRENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPA 90
G Y + P G + Q+E+ PERPDQ ECQ YMKTGDCKFG+ CR+HHP + + A
Sbjct: 404 GPYQSSGPSTGPSG--SSQKEHSLPERPDQQECQHYMKTGDCKFGSTCRYHHPPD--MGA 571
Query: 91 PDCVLSPLGLPLRPG 105
P LSP+GLPLRPG
Sbjct: 572 PKVNLSPIGLPLRPG 616
Score = 41.2 bits (95), Expect = 3e-04
Identities = 17/43 (39%), Positives = 25/43 (57%)
Frame = +2
Query: 100 LPLRPGEPLCVFYSRYGICKFGPSCKFDHPMGIFTYNVSASPL 142
LP RP + C Y + G CKFG +C++ HP + V+ SP+
Sbjct: 467 LPERPDQQECQHYMKTGDCKFGSTCRYHHPPDMGAPKVNLSPI 595
Score = 26.6 bits (57), Expect = 6.7
Identities = 9/20 (45%), Positives = 12/20 (60%)
Frame = +1
Query: 105 GEPLCVFYSRYGICKFGPSC 124
G C Y++ G CKFG +C
Sbjct: 799 GAQPCTHYTQRGFCKFGSAC 858
>TC84075 similar to GP|3738297|gb|AAC63639.1| unknown protein {Arabidopsis
thaliana}, partial (17%)
Length = 615
Score = 83.6 bits (205), Expect = 5e-17
Identities = 42/89 (47%), Positives = 53/89 (59%)
Frame = +3
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYS 113
+PER P C +YM+TG C +G CRF+HPR+R A V + P R GEP C +Y
Sbjct: 324 YPERHGVPNCAYYMRTGFCGYGGRCRFNHPRDRAAVAA-AVRATGDYPERLGEPPCQYYL 500
Query: 114 RYGICKFGPSCKFDHPMGIFTYNVSASPL 142
+ G CKFG SCKF HP Y +S +PL
Sbjct: 501 KTGTCKFGASCKFHHPKNGGGY-LSQAPL 584
Score = 64.7 bits (156), Expect = 2e-11
Identities = 28/59 (47%), Positives = 37/59 (62%)
Frame = +3
Query: 47 ALQRENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPG 105
A++ +PER +P CQ+Y+KTG CKFGA C+FHHP+ L+ G PLRPG
Sbjct: 438 AVRATGDYPERLGEPPCQYYLKTGTCKFGASCKFHHPKNGGGYLSQAPLNIYGYPLRPG 614
>AL365712 similar to GP|3738297|gb|A unknown protein {Arabidopsis thaliana},
partial (5%)
Length = 520
Score = 69.3 bits (168), Expect = 9e-13
Identities = 28/45 (62%), Positives = 35/45 (77%)
Frame = +1
Query: 60 QPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRP 104
+P CQ+YM+TGDCKFG CR+HHPR++ P +LSP GLPLRP
Sbjct: 4 EPVCQYYMRTGDCKFGLACRYHHPRDQVAARP--LLSPFGLPLRP 132
Score = 42.0 bits (97), Expect = 2e-04
Identities = 19/43 (44%), Positives = 25/43 (57%)
Frame = +1
Query: 105 GEPLCVFYSRYGICKFGPSCKFDHPMGIFTYNVSASPLAEAAG 147
GEP+C +Y R G CKFG +C++ HP V+A PL G
Sbjct: 1 GEPVCQYYMRTGDCKFGLACRYHHPRD----QVAARPLLSPFG 117
>TC79991 similar to GP|10177307|dbj|BAB10568. contains similarity to zinc
finger protein~gene_id:MDC12.23 {Arabidopsis thaliana},
partial (8%)
Length = 1023
Score = 52.0 bits (123), Expect = 1e-07
Identities = 20/33 (60%), Positives = 24/33 (72%)
Frame = +1
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRER 86
FP RP+ +C FYMKTG CKFG C+F+HP R
Sbjct: 856 FPLRPEAEDCSFYMKTGSCKFGFNCKFNHPIRR 954
Score = 42.4 bits (98), Expect = 1e-04
Identities = 17/30 (56%), Positives = 21/30 (69%)
Frame = +1
Query: 101 PLRPGEPLCVFYSRYGICKFGPSCKFDHPM 130
PLRP C FY + G CKFG +CKF+HP+
Sbjct: 859 PLRPEAEDCSFYMKTGSCKFGFNCKFNHPI 948
>TC83740 similar to GP|6437560|gb|AAF08587.1| hypothetical protein
{Arabidopsis thaliana}, partial (10%)
Length = 435
Score = 52.0 bits (123), Expect = 1e-07
Identities = 17/33 (51%), Positives = 28/33 (84%)
Frame = +2
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRER 86
+P+RPD+ +C +Y++TG C +G+ CRF+HPR+R
Sbjct: 296 YPQRPDEADCIYYLRTGFCGYGSRCRFNHPRDR 394
Score = 40.0 bits (92), Expect = 6e-04
Identities = 14/29 (48%), Positives = 19/29 (65%)
Frame = +2
Query: 101 PLRPGEPLCVFYSRYGICKFGPSCKFDHP 129
P RP E C++Y R G C +G C+F+HP
Sbjct: 299 PQRPDEADCIYYLRTGFCGYGSRCRFNHP 385
>TC82244 similar to GP|22507458|gb|AAH19429.1 Unknown (protein for
MGC:30371) {Mus musculus}, partial (13%)
Length = 847
Score = 44.3 bits (103), Expect = 3e-05
Identities = 16/31 (51%), Positives = 21/31 (67%)
Frame = +3
Query: 53 IFPERPDQPECQFYMKTGDCKFGAVCRFHHP 83
I+P+RP + +C YM T CKFG C+F HP
Sbjct: 675 IYPQRPGEKDCAHYMLTRTCKFGESCKFDHP 767
Score = 43.9 bits (102), Expect = 4e-05
Identities = 18/32 (56%), Positives = 20/32 (62%)
Frame = +3
Query: 101 PLRPGEPLCVFYSRYGICKFGPSCKFDHPMGI 132
P RPGE C Y CKFG SCKFDHP+ +
Sbjct: 681 PQRPGEKDCAHYMLTRTCKFGESCKFDHPIWV 776
>TC81206 similar to GP|8777433|dbj|BAA97023.1 gene_id:MHM17.4~unknown
protein {Arabidopsis thaliana}, partial (5%)
Length = 842
Score = 39.3 bits (90), Expect = 0.001
Identities = 27/92 (29%), Positives = 40/92 (43%), Gaps = 1/92 (1%)
Frame = +3
Query: 48 LQRENIFPERPDQP-ECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGE 106
++R + PE+ +P +C+ YM G C G C F H +PL +
Sbjct: 69 VKRPKLDPEQKPKPKQCRHYMN-GRCHKGDECNFSHD---------------AIPLTKSK 200
Query: 107 PLCVFYSRYGICKFGPSCKFDHPMGIFTYNVS 138
P CV Y+ + C G C +DH +F Y S
Sbjct: 201 P-CVHYAHHRSCMKGNDCPYDH--DLFNYPCS 287
>TC81324 similar to GP|21553871|gb|AAM62964.1 unknown {Arabidopsis
thaliana}, partial (75%)
Length = 1156
Score = 36.6 bits (83), Expect = 0.006
Identities = 24/87 (27%), Positives = 30/87 (33%), Gaps = 21/87 (24%)
Frame = +1
Query: 63 CQFYMKTGDCKFGAVCRFHH--------------------PRERTIPAPDCVLSPLG-LP 101
C + T C FG C F H P R +PAP +P G P
Sbjct: 208 CTKFFSTSGCPFGESCHFLHHVPGGYNAVSQMMNLTPAAPPAPRNVPAPRNAHAPNGSAP 387
Query: 102 LRPGEPLCVFYSRYGICKFGPSCKFDH 128
+C ++ CKFG C F H
Sbjct: 388 SAVKSRICSKFNTAEGCKFGDKCHFAH 468
>BE998796 similar to GP|5441893|dbj| ESTs C99174(E10437) D22295(C10709)
correspond to a region of the predicted gene.~Similar
to, partial (28%)
Length = 609
Score = 33.5 bits (75), Expect = 0.055
Identities = 23/81 (28%), Positives = 28/81 (34%), Gaps = 21/81 (25%)
Frame = +1
Query: 69 TGDCKFGAVCRFHH--------------------PRERTIPAPDCVLSPLG-LPLRPGEP 107
T C FG C F H P R +PAP +P G P
Sbjct: 169 TSGCPFGESCHFLHHVPGGYNAVSQMMNLTPAAPPAPRNVPAPRNAHAPNGSAPSAVKSR 348
Query: 108 LCVFYSRYGICKFGPSCKFDH 128
+C ++ CKFG C F H
Sbjct: 349 ICSKFNTAEGCKFGDKCHFAH 411
>TC92368 weakly similar to GP|19347974|gb|AAL86319.1 unknown protein
{Arabidopsis thaliana}, partial (63%)
Length = 999
Score = 33.1 bits (74), Expect = 0.072
Identities = 17/68 (25%), Positives = 28/68 (41%)
Frame = +3
Query: 63 CQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSRYGICKFGP 122
C ++ +G C++G C F H T C+ +C + G C+ GP
Sbjct: 93 CFKFVSSGSCQWGETCNFRHD---TDAREHCL-----------RGVCFDFLNKGKCERGP 230
Query: 123 SCKFDHPM 130
C+F H +
Sbjct: 231 DCRFRHSL 254
Score = 26.2 bits (56), Expect = 8.8
Identities = 9/24 (37%), Positives = 14/24 (57%)
Frame = +3
Query: 105 GEPLCVFYSRYGICKFGPSCKFDH 128
G+ LC + G C++G +C F H
Sbjct: 81 GDKLCFKFVSSGSCQWGETCNFRH 152
>TC81337 similar to GP|15723293|gb|AAL06332.1 U2 auxiliary factor small
subunit {Arabidopsis thaliana}, partial (81%)
Length = 1190
Score = 32.0 bits (71), Expect = 0.16
Identities = 20/65 (30%), Positives = 26/65 (39%), Gaps = 12/65 (18%)
Frame = +2
Query: 52 NIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPA---------PDCV---LSPLG 99
+IF D+ C FY K G C+ G C H R P PD + + P G
Sbjct: 101 SIFGTEKDRVNCPFYFKIGACRHGDRCSRLHNRPTISPTLLLSNMYQRPDIITPGVDPNG 280
Query: 100 LPLRP 104
P+ P
Sbjct: 281 QPIDP 295
>AL366906 weakly similar to PIR|T45743|T45 hypothetical protein F24M12.160 -
Arabidopsis thaliana, partial (2%)
Length = 486
Score = 31.2 bits (69), Expect = 0.27
Identities = 13/29 (44%), Positives = 18/29 (61%)
Frame = +3
Query: 101 PLRPGEPLCVFYSRYGICKFGPSCKFDHP 129
PL G+ +C FY G C+ G SC++ HP
Sbjct: 258 PLPKGQRVCKFYES-GYCRKGASCEYFHP 341
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.137 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,949,612
Number of Sequences: 36976
Number of extensions: 128065
Number of successful extensions: 647
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 561
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 611
length of query: 197
length of database: 9,014,727
effective HSP length: 91
effective length of query: 106
effective length of database: 5,649,911
effective search space: 598890566
effective search space used: 598890566
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC148289.7


