

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148289.2 + phase: 0 /pseudo
(440 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
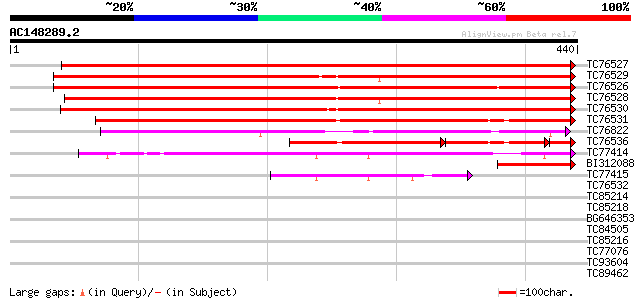
Score E
Sequences producing significant alignments: (bits) Value
TC76527 similar to PIR|S35757|S35757 vicilin 47K - garden pea, ... 784 0.0
TC76529 similar to PIR|S35757|S35757 vicilin 47K - garden pea, ... 719 0.0
TC76526 similar to SP|P13918|VCLC_PEA Vicilin precursor. [Garden... 694 0.0
TC76528 similar to PIR|S35757|S35757 vicilin 47K - garden pea, ... 693 0.0
TC76530 similar to SP|P13918|VCLC_PEA Vicilin precursor. [Garden... 679 0.0
TC76531 similar to SP|P13915|CVCA_PEA Convicilin precursor. [Gar... 527 e-150
TC76822 similar to PIR|T06459|T06459 62K sucrose-binding protein... 195 3e-50
TC76536 similar to GP|1297070|emb|CAA96513.1 convicilin precurso... 167 1e-41
TC77414 similar to GP|12697782|dbj|BAB21619. allergen Gly m Bd 2... 120 1e-27
BI312088 similar to PIR|T47865|T4786 transport protein subunit-l... 109 2e-24
TC77415 similar to GP|12697782|dbj|BAB21619. allergen Gly m Bd 2... 57 2e-08
TC76532 weakly similar to SP|Q9BBN6|YCF1_LOTJA Hypothetical 214.... 41 0.001
TC85214 similar to PIR|S26688|S26688 legumin K - garden pea, par... 37 0.015
TC85218 similar to SP|P05692|LEGJ_PEA Legumin J precursor. [Gard... 35 0.057
BG646353 weakly similar to GP|12697782|db allergen Gly m Bd 28K ... 34 0.098
TC84505 similar to PIR|T48442|T48442 hypothetical protein T32M21... 33 0.17
TC85216 similar to PIR|S26688|S26688 legumin K - garden pea, par... 33 0.29
TC77076 similar to PIR|T07734|T07734 homeotic protein VAHOX1 - t... 32 0.64
TC93604 weakly similar to SP|Q9SR72|GL32_ARATH Germin-like prote... 32 0.64
TC89462 similar to GP|9294563|dbj|BAB02826.1 gene_id:MQC12.22~un... 31 0.83
>TC76527 similar to PIR|S35757|S35757 vicilin 47K - garden pea, partial
(96%)
Length = 1537
Score = 784 bits (2024), Expect = 0.0
Identities = 399/399 (100%), Positives = 399/399 (100%)
Frame = +1
Query: 41 INMAIKAPCSLLMLLGIVFLASICVSSRSDQDQENPFIFNSNRFQTLFENENGHIRLLQR 100
INMAIKAPCSLLMLLGIVFLASICVSSRSDQDQENPFIFNSNRFQTLFENENGHIRLLQR
Sbjct: 1 INMAIKAPCSLLMLLGIVFLASICVSSRSDQDQENPFIFNSNRFQTLFENENGHIRLLQR 180
Query: 101 FDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNL 160
FDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNL
Sbjct: 181 FDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNL 360
Query: 161 ERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSK 220
ERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSK
Sbjct: 361 ERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSK 540
Query: 221 NILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSK 280
NILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSK
Sbjct: 541 NILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSK 720
Query: 281 NAKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGS 340
NAKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGS
Sbjct: 721 NAKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGS 900
Query: 341 LLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLS 400
LLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLS
Sbjct: 901 LLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLS 1080
Query: 401 PGDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
PGDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA
Sbjct: 1081PGDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 1197
>TC76529 similar to PIR|S35757|S35757 vicilin 47K - garden pea, partial
(94%)
Length = 1588
Score = 719 bits (1855), Expect = 0.0
Identities = 372/415 (89%), Positives = 389/415 (93%), Gaps = 10/415 (2%)
Frame = +1
Query: 35 SEIISKINMAIKAPCSLLMLLGIVFLASICVSSRSDQDQENPFIFNSNRFQTLFENENGH 94
S+IISKINMAIKAP SLLMLLG VFLAS+CVSSRSDQDQENPFIFNSNRFQTLFENENGH
Sbjct: 4 SDIISKINMAIKAPSSLLMLLGTVFLASVCVSSRSDQDQENPFIFNSNRFQTLFENENGH 183
Query: 95 IRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNN 154
IRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFIL V+SGKAILTVLNPNN
Sbjct: 184 IRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILAVLSGKAILTVLNPNN 363
Query: 155 RNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSF 214
RNSFNLERGDTIKLPAG++ YLANRDDN+DLRVLDLAIPVNRPGQFQSFSLS ++NQQSF
Sbjct: 364 RNSFNLERGDTIKLPAGSIAYLANRDDNQDLRVLDLAIPVNRPGQFQSFSLSGNQNQQSF 543
Query: 215 LSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQ 274
SGFSKNILEAAFN+NYEEIERVLIE EQEPQHRRGLR D R +QSQEA+VIVKVSREQ
Sbjct: 544 FSGFSKNILEAAFNANYEEIERVLIE--EQEPQHRRGLR-DRRHKQSQEADVIVKVSREQ 714
Query: 275 IEELSKNAKSS----------SRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQL 324
IEELS++AKSS SRRS SSES P NLR+ +PIYSN+FGNFFEITPEKNPQL
Sbjct: 715 IEELSRHAKSSSRRSASSESASRRSASSESAPFNLRSHEPIYSNEFGNFFEITPEKNPQL 894
Query: 325 KDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQ 384
+DLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQ
Sbjct: 895 QDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQ 1074
Query: 385 QQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
QQERSQQVQRYRARLSPGDVYVIPAGHPIVV ASSDLSLLGFGINAENNQRNFLA
Sbjct: 1075QQERSQQVQRYRARLSPGDVYVIPAGHPIVVKASSDLSLLGFGINAENNQRNFLA 1239
>TC76526 similar to SP|P13918|VCLC_PEA Vicilin precursor. [Garden pea] {Pisum
sativum}, partial (95%)
Length = 2210
Score = 694 bits (1792), Expect = 0.0
Identities = 354/406 (87%), Positives = 384/406 (94%), Gaps = 1/406 (0%)
Frame = +2
Query: 35 SEIISKINMAIKAPCSLLMLLGIVFLASICVSSRSDQ-DQENPFIFNSNRFQTLFENENG 93
S + +I MAIKAP LLMLLGI FLAS+CVSSR D+ DQENPF FN+N FQTLFENENG
Sbjct: 659 SVFLIQIIMAIKAPFQLLMLLGIFFLASVCVSSRDDRHDQENPFFFNANHFQTLFENENG 838
Query: 94 HIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPN 153
HIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFIL V+SGKAILTVLNP+
Sbjct: 839 HIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILAVLSGKAILTVLNPD 1018
Query: 154 NRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQS 213
NRNSFNLERGDTIKLPAG++ YLANRDDN+DLRVLDLAIPVNRPG+FQSFSLS S+NQQS
Sbjct: 1019 NRNSFNLERGDTIKLPAGSIAYLANRDDNEDLRVLDLAIPVNRPGKFQSFSLSGSQNQQS 1198
Query: 214 FLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSRE 273
F SGFSKNILEAAFN+NYEEIERVLIEE+EQEPQHRRGLRKD RRQQSQ++NVIVKVSRE
Sbjct: 1199 FFSGFSKNILEAAFNANYEEIERVLIEEHEQEPQHRRGLRKD-RRQQSQDSNVIVKVSRE 1375
Query: 274 QIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNY 333
QIEELS++AKSSSRRS SSES P NLR+++PIYSN+FGNFFEITPEKNPQLKDLDILVNY
Sbjct: 1376 QIEELSRHAKSSSRRSGSSESAPFNLRSREPIYSNEFGNFFEITPEKNPQLKDLDILVNY 1555
Query: 334 AEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQ 393
AEIREGSLLLPHFNSRATVIV V+EGKGEFELVGQRNENQQEQRE E+++Q++ERSQQVQ
Sbjct: 1556 AEIREGSLLLPHFNSRATVIVVVDEGKGEFELVGQRNENQQEQRE-EDEQQEEERSQQVQ 1732
Query: 394 RYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
RYRARLSPGDVYVIPAGHP VV+ASSDLSLLGFGINAENN+RNFLA
Sbjct: 1733 RYRARLSPGDVYVIPAGHPTVVSASSDLSLLGFGINAENNERNFLA 1870
>TC76528 similar to PIR|S35757|S35757 vicilin 47K - garden pea, partial
(95%)
Length = 1566
Score = 693 bits (1788), Expect = 0.0
Identities = 355/407 (87%), Positives = 378/407 (92%), Gaps = 10/407 (2%)
Frame = +2
Query: 43 MAIKAPCSLLMLLGIVFLASICVSSRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFD 102
MAIKAP LLMLLGI FLAS+CVSSRSD DQENPF FN+NRFQTLFENENGHIRLLQRFD
Sbjct: 20 MAIKAPFQLLMLLGIFFLASVCVSSRSDHDQENPFFFNANRFQTLFENENGHIRLLQRFD 199
Query: 103 KRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLER 162
KRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFIL V+SGKAILTVLNPN+RNSFNLER
Sbjct: 200 KRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILAVLSGKAILTVLNPNDRNSFNLER 379
Query: 163 GDTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNI 222
GDTIKLPAG++ YLANR DN+DLRVLDLAIPVNRPGQFQSFSLS ++NQQSF SGFSKNI
Sbjct: 380 GDTIKLPAGSIAYLANRADNEDLRVLDLAIPVNRPGQFQSFSLSGNQNQQSFFSGFSKNI 559
Query: 223 LEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNA 282
LEAAFNSNYEEIERVLIEE EQEP+HRRGLR D R +QSQEANVIVKVSREQIEELS++A
Sbjct: 560 LEAAFNSNYEEIERVLIEEQEQEPRHRRGLR-DRRHKQSQEANVIVKVSREQIEELSRHA 736
Query: 283 KSS----------SRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVN 332
KSS SRRS SSES P NLR+++PIYSN+FGNFFEITPEKNPQL+DLDILVN
Sbjct: 737 KSSSRRSASSESASRRSASSESAPFNLRSREPIYSNEFGNFFEITPEKNPQLQDLDILVN 916
Query: 333 YAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQV 392
YAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQ++ERSQQV
Sbjct: 917 YAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQV 1096
Query: 393 QRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
QRYRARL+PGDVYVIPAG+P VV ASSDLSLLGFGINAENNQR+FLA
Sbjct: 1097QRYRARLTPGDVYVIPAGYPNVVKASSDLSLLGFGINAENNQRSFLA 1237
>TC76530 similar to SP|P13918|VCLC_PEA Vicilin precursor. [Garden pea]
{Pisum sativum}, partial (90%)
Length = 1549
Score = 679 bits (1752), Expect = 0.0
Identities = 351/402 (87%), Positives = 375/402 (92%), Gaps = 2/402 (0%)
Frame = +2
Query: 40 KINMAIKAPCSLLMLLGIVFLASICVSSRSDQ-DQENPFIFNSNRFQTLFENENGHIRLL 98
+I MAIKAP L MLLGI FLAS+CVSS+ DQ DQENPF FN+NRFQTLFENENGHIRLL
Sbjct: 5 QIIMAIKAPFQLFMLLGIFFLASVCVSSKDDQHDQENPFFFNANRFQTLFENENGHIRLL 184
Query: 99 QRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSF 158
QRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFIL V+SGKAILTVLNPNNRNSF
Sbjct: 185 QRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILAVLSGKAILTVLNPNNRNSF 364
Query: 159 NLERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGF 218
NLERGDTIKLPAG++ YLANRDDN+DLRVLDLAIPVNRPGQFQSFSLS S+NQQSF SGF
Sbjct: 365 NLERGDTIKLPAGSIAYLANRDDNQDLRVLDLAIPVNRPGQFQSFSLSGSQNQQSFFSGF 544
Query: 219 SKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEEL 278
SKNILEAAFN+NYEEIERVLIEE EQ+P HRRGLR D R +QSQEANVIVKVSREQIEEL
Sbjct: 545 SKNILEAAFNANYEEIERVLIEEQEQDP-HRRGLR-DRRHKQSQEANVIVKVSREQIEEL 718
Query: 279 SKNAK-SSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIR 337
S++AK SSSRRS SSES P NLR++KPIYSN+FGNFFEITPEKN QL+DLDILV AEIR
Sbjct: 719 SRHAKSSSSRRSASSESAPFNLRSRKPIYSNEFGNFFEITPEKNQQLQDLDILVTNAEIR 898
Query: 338 EGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRA 397
EGSLLLPHFNSRATVIV V+EGKGEFELVGQRNENQQEQREYEEDEQ++ERSQQVQRYRA
Sbjct: 899 EGSLLLPHFNSRATVIVVVDEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQVQRYRA 1078
Query: 398 RLSPGDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
RL+PGDVYVIPAG+P VV ASSDLSLLGFGINAENNQR+FLA
Sbjct: 1079RLTPGDVYVIPAGYPNVVKASSDLSLLGFGINAENNQRSFLA 1204
>TC76531 similar to SP|P13915|CVCA_PEA Convicilin precursor. [Garden pea]
{Pisum sativum}, partial (86%)
Length = 2217
Score = 527 bits (1357), Expect = e-150
Identities = 262/373 (70%), Positives = 314/373 (83%)
Frame = +2
Query: 67 SRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTL 126
S + + NPF+F SNRFQTLFENENGHIRL+QRFDKRS +F+NL+NYRLLEY +KPHT+
Sbjct: 728 SSESEGRRNPFLFRSNRFQTLFENENGHIRLIQRFDKRSNLFQNLKNYRLLEYRAKPHTI 907
Query: 127 FLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLR 186
FLP H DADFILVV+SGKAILTVLN NNRNSFNLE+GDTIKLPAG+ YL N+D+++DLR
Sbjct: 908 FLPHHTDADFILVVLSGKAILTVLNSNNRNSFNLEQGDTIKLPAGSTAYLVNQDNDEDLR 1087
Query: 187 VLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEP 246
V+DL IPVNRPG+FQSF LS ++N S+ GFSK+ILEA+FN++YE IERVL+EE+E E
Sbjct: 1088 VVDLVIPVNRPGKFQSFDLSGNQNNPSYFRGFSKSILEASFNTDYETIERVLLEEHEHEQ 1267
Query: 247 QHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIY 306
Q RRG + RQQSQEAN IVKVSREQIEEL ++AKSSS+RS SSES P NLRN+ P+Y
Sbjct: 1268 QQRRGRK---GRQQSQEANAIVKVSREQIEELRRHAKSSSKRSISSESGPFNLRNRNPLY 1438
Query: 307 SNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELV 366
SNKFG FFEITPEKNPQL+DLDI V+ EI EG+L+LPH+NSR+TV++ + EGKG ELV
Sbjct: 1439 SNKFGKFFEITPEKNPQLQDLDIFVSSVEINEGALILPHYNSRSTVVLVMNEGKGHLELV 1618
Query: 367 GQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGF 426
G ++E QQEQRE +E E R+ +VQRY ARLSPGDV +IPAGHP+ + ASS+L+ LGF
Sbjct: 1619 GHKSE-QQEQREQDEKEL---RNPEVQRYNARLSPGDVVIIPAGHPVAINASSNLNFLGF 1786
Query: 427 GINAENNQRNFLA 439
GINAENNQRNFLA
Sbjct: 1787 GINAENNQRNFLA 1825
Score = 32.3 bits (72), Expect = 0.37
Identities = 16/34 (47%), Positives = 24/34 (70%), Gaps = 2/34 (5%)
Frame = +2
Query: 45 IKAPCSLLMLLGIVFLASICVSSR--SDQDQENP 76
IK+ LL+LLGI+FL S+CVS +Q++ +P
Sbjct: 32 IKSRFPLLLLLGIIFLGSVCVSYGIVGEQEERHP 133
>TC76822 similar to PIR|T06459|T06459 62K sucrose-binding protein homolog -
garden pea, partial (97%)
Length = 1693
Score = 195 bits (495), Expect = 3e-50
Identities = 115/371 (30%), Positives = 192/371 (50%), Gaps = 6/371 (1%)
Frame = +2
Query: 71 QDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQ 130
++ ENP++F F+T + ++G + L FD++SK+ N +NY L +K H P
Sbjct: 287 EEDENPYVFEDRDFETKIDTDDGRVMALNMFDQKSKLLRNFENYGLTILEAKGHAFVSPH 466
Query: 131 HNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVLDL 190
H D++ I V G+ I+ ++ + FNLE GD +++PAGT YL NRD+N+ L V L
Sbjct: 467 HFDSEVIFFNVKGRGIIGLVMEDKTERFNLEAGDIMRVPAGTPMYLVNRDENEKLFVAAL 646
Query: 191 AIP---VNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQ 247
+P + P ++F + +S L+ FS +L+AAF S ++E L E+N+
Sbjct: 647 HMPPSSASAPVNLEAFFGPAGRDPESVLTAFSSKVLQAAFKSPKGKLESFLDEQNKGR-- 820
Query: 248 HRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYS 307
I K+ +E + L+ K S P N+ + P +S
Sbjct: 821 -------------------IFKIQKEDLSGLA--PKKSIWPFGGQFKNPFNIFSNNPAFS 937
Query: 308 NKFGNFFEITP-EKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELV 366
N+FG+ FE+ P E L L++++ +A I +GS+ ++N+ A I V +G+GEFE+
Sbjct: 938 NQFGSLFEVGPSEITSGLDGLNLMLTFANITKGSMSTIYYNTNANKIALVIDGEGEFEMA 1117
Query: 367 GQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTAS--SDLSLL 424
+ +Q++ S Q+ ARL PG V+V+PAGHP V AS ++L ++
Sbjct: 1118CPHMSSSS-----SHSKQRRSSSTSYQKINARLRPGTVFVVPAGHPFVTIASKNNNLKIV 1282
Query: 425 GFGINAENNQR 435
F +NA+ N++
Sbjct: 1283CFEVNAQRNKK 1315
>TC76536 similar to GP|1297070|emb|CAA96513.1 convicilin precursor {Vicia
narbonensis}, partial (33%)
Length = 878
Score = 167 bits (422), Expect = 1e-41
Identities = 87/121 (71%), Positives = 101/121 (82%)
Frame = +2
Query: 218 FSKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEE 277
FSK+ILEA+FN++YE IERVL+EE+E E Q RRG + RQQSQEAN IVKVSREQIEE
Sbjct: 20 FSKSILEASFNTDYETIERVLLEEHEHEQQQRRGRKG---RQQSQEANAIVKVSREQIEE 190
Query: 278 LSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIR 337
L ++AKSSS+RS SSES P NLRN+ P+YSNKFG FFEITPEKNPQL+DLDI V+ EI
Sbjct: 191 LRRHAKSSSKRSISSESGPFNLRNRNPLYSNKFGKFFEITPEKNPQLQDLDIFVSSVEIN 370
Query: 338 E 338
E
Sbjct: 371 E 373
Score = 100 bits (250), Expect(2) = 2e-25
Identities = 49/81 (60%), Positives = 64/81 (78%)
Frame = +1
Query: 339 GSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRAR 398
G+L+LPH+NSR+TV++ + EGKG ELVG ++E QQEQRE +E E R+ +VQRY AR
Sbjct: 571 GALILPHYNSRSTVVLVMNEGKGHLELVGHKSE-QQEQREQDEKEL---RNPEVQRYNAR 738
Query: 399 LSPGDVYVIPAGHPIVVTASS 419
LSPGDV +IPAGHP+ + ASS
Sbjct: 739 LSPGDVVIIPAGHPVAINASS 801
Score = 32.7 bits (73), Expect(2) = 2e-25
Identities = 15/20 (75%), Positives = 17/20 (85%)
Frame = +2
Query: 420 DLSLLGFGINAENNQRNFLA 439
+L LGFGINAE +QRNFLA
Sbjct: 803 NLISLGFGINAEKHQRNFLA 862
>TC77414 similar to GP|12697782|dbj|BAB21619. allergen Gly m Bd 28K {Glycine
max}, partial (79%)
Length = 1575
Score = 120 bits (301), Expect = 1e-27
Identities = 103/418 (24%), Positives = 177/418 (41%), Gaps = 32/418 (7%)
Frame = +3
Query: 54 LLGIVFLASICVSSRSDQDQE-----NPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIF 108
LL + L + ++ D+++E N F+ +++ ++ + G +RL + D R
Sbjct: 12 LLFFLLLGGVTITMCEDEEREEGSSSNLFLMQNSK--SVVKTHAGELRLFKNNDDR--FL 179
Query: 109 ENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKL 168
+ + LL + +P +LF+PQ+ D++ I+ V G A L + + ++ GD +
Sbjct: 180 DRHMHIGLL--NMEPRSLFIPQYLDSNLIIFVRRGVAKLGFIYGDELEERRIKTGDLYVI 353
Query: 169 PAGTLGYLANRDDNKDLRVLDLAIPVNRPGQ-FQSFSLSESENQQSFLSGFSKNILEAAF 227
PAGT+ YL N + + L V+ P G FQ F + +NQQS L+GF ILE AF
Sbjct: 354 PAGTVFYLVNIGEGQRLHVICSFDPSTSLGDTFQPFYIGGGDNQQSVLAGFGPTILETAF 533
Query: 228 NSNYEEIER----------VLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEE 277
N + +IER V I+++ P + ++ + Q +V+ E+ EE
Sbjct: 534 NESRTKIERIFTKKQDGPIVFIDDDSHSPSLWTKFLELKKNDKVQHLKTLVQRQEEEEEE 713
Query: 278 --------------LSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQ 323
L K K + + + NL ++KP + N +G +
Sbjct: 714 KQTSWSWRKLMKNVLGKEKKKIENKDRADSPDSYNLYDRKPDFRNAYGWSSALDGGDYSP 893
Query: 324 LKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDE 383
LK DI V + + GS++ PH N AT V G G +++ N E
Sbjct: 894 LKIPDIGVFHVNLTAGSMMAPHVNPSATEYTIVLRGYGRIQILFPNGSNAME-------- 1049
Query: 384 QQQERSQQVQRYRARLSPGDVYVIPAGHPI--VVTASSDLSLLGFGINAENNQRNFLA 439
A + GD++ IP P + + L GF +++ + FLA
Sbjct: 1050-------------AEIKVGDIFYIPRYFPFCQIAARNGPLEFFGFTTSSKKSYPQFLA 1184
>BI312088 similar to PIR|T47865|T4786 transport protein subunit-like -
Arabidopsis thaliana, partial (80%)
Length = 663
Score = 109 bits (272), Expect = 2e-24
Identities = 53/61 (86%), Positives = 59/61 (95%)
Frame = +1
Query: 379 YEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFL 438
+EEDEQ++ERSQQVQRYRARL+PGDVYVIPAG+P VV ASSDLSLLGFGINAENNQR+FL
Sbjct: 475 HEEDEQEEERSQQVQRYRARLTPGDVYVIPAGYPNVVKASSDLSLLGFGINAENNQRSFL 654
Query: 439 A 439
A
Sbjct: 655 A 657
>TC77415 similar to GP|12697782|dbj|BAB21619. allergen Gly m Bd 28K {Glycine
max}, partial (51%)
Length = 978
Score = 56.6 bits (135), Expect = 2e-08
Identities = 46/186 (24%), Positives = 76/186 (40%), Gaps = 29/186 (15%)
Frame = +1
Query: 203 FSLSESENQQSFLSGFSKNILEAAFNSNYEEIER----------VLIEENEQEPQHRRGL 252
F + +NQQS L+GF ILE AFN + +IER V I+++ P
Sbjct: 427 FYIGGGDNQQSVLAGFGPTILETAFNESRTKIERIFTKKQDGPIVFIDDDSHSPSLWTKF 606
Query: 253 RKDERRQQSQEANVIVKVSREQIEE--------------LSKNAKSSSRRSESSESEPIN 298
+ ++ + Q +V+ E+ EE L K K + + + N
Sbjct: 607 LELKKNDKVQHLKTLVQRQEEEEEEKQTSWSWRKLMKNVLGKEKKKIENKDRADSPDSYN 786
Query: 299 LRNQKPIYSNKFG-----NFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVI 353
L ++KP + N +G ++ + + P DI V + + G ++ PH N AT
Sbjct: 787 LYDRKPDFRNAYGLEQCIGWWRLFSAQIP-----DIGVFHVNLTRGIMMAPHVNPSATEY 951
Query: 354 VAVEEG 359
V G
Sbjct: 952 TIVLRG 969
>TC76532 weakly similar to SP|Q9BBN6|YCF1_LOTJA Hypothetical 214.8 kDa
protein ycf1. {Lotus japonicus}, partial (21%)
Length = 4509
Score = 40.8 bits (94), Expect = 0.001
Identities = 20/25 (80%), Positives = 22/25 (88%)
Frame = -1
Query: 255 DERRQQSQEANVIVKVSREQIEELS 279
+ +QSQEANVIVKVSREQIEELS
Sbjct: 75 NRHEEQSQEANVIVKVSREQIEELS 1
>TC85214 similar to PIR|S26688|S26688 legumin K - garden pea, partial (94%)
Length = 1831
Score = 37.0 bits (84), Expect = 0.015
Identities = 25/129 (19%), Positives = 53/129 (40%), Gaps = 2/129 (1%)
Frame = +3
Query: 240 EENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINL 299
EE E++P+H R +++ R+ +E E+ ++ + S N+
Sbjct: 1029 EERERDPRHPRHSQEEREREDDPYGRGRPWWEKESREKQRTRGQNGLEETICSARLVENI 1208
Query: 300 RN--QKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVE 357
+Y+ + G ++ P L++L + Y + + PH+N A ++ V
Sbjct: 1209 ARPAHADLYNPRAGRISDVNSLTLPILRNLRLSAEYVLLYRNGIYAPHWNINANSLLYVI 1388
Query: 358 EGKGEFELV 366
G+G +V
Sbjct: 1389 RGQGRVRIV 1415
Score = 34.7 bits (78), Expect = 0.075
Identities = 27/102 (26%), Positives = 47/102 (45%), Gaps = 10/102 (9%)
Frame = +3
Query: 321 NPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELV----------GQRN 370
+P+L+ + + I L LP ++ +I ++ GKG L Q +
Sbjct: 174 HPELQCAGVSLIRRTIDPNGLHLPSYSPSPQLIFIIQ-GKGVLGLSVPGCPETFEQPQSS 350
Query: 371 ENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHP 412
++Q R E+ +QQ + Q+++R+ GDV IPAG P
Sbjct: 351 RSRQGSRHQEQQQQQPDSHQKIRRFYR----GDVIAIPAGTP 464
>TC85218 similar to SP|P05692|LEGJ_PEA Legumin J precursor. [Garden pea]
{Pisum sativum}, partial (73%)
Length = 1595
Score = 35.0 bits (79), Expect = 0.057
Identities = 46/245 (18%), Positives = 84/245 (33%), Gaps = 69/245 (28%)
Frame = +2
Query: 191 AIPVNRPGQFQSFSLSESENQ-QSFLSGFSKNILEAAFNSNYEEIER-----------VL 238
+ P R G+ Q SE +N+ S LSGFS L A N++ + +R V
Sbjct: 371 SFPGRRGGRQQQEEGSEEQNEGSSVLSGFSSEFLAQALNTDQDTAKRLQSPRDQRSQIVR 550
Query: 239 IE---------------------ENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEE 277
+E E E++ + R +R+ ++ S+E + R EE
Sbjct: 551 VEGGLSIISPEWQQEDEEYERSHEEEEDERRPRHIRRPGHQKPSEEEQWETRYPRHSQEE 730
Query: 278 LSKNAKSSSRRSESSESEPINL------RNQKP--------------------------- 304
++ + + E +P R + P
Sbjct: 731 RERDPRRPGHSQKEREWDPRRPGHGQEERERDPRHSGHSQNGLEETICSLRIVENIARPA 910
Query: 305 ---IYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKG 361
+Y+ + G + P L++L + Y + + PH+N A ++ V G+G
Sbjct: 911 RADLYNPRAGRISDANSLTLPILRNLRLSAEYVLLYRNGIYAPHWNINANSLLYVIRGQG 1090
Query: 362 EFELV 366
+V
Sbjct: 1091RVRIV 1105
Score = 32.0 bits (71), Expect = 0.49
Identities = 14/44 (31%), Positives = 26/44 (58%)
Frame = +2
Query: 369 RNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHP 412
R+ +++ ++E +QQ + Q+++R+ GDV IPAG P
Sbjct: 77 RSSRSRQESRHQEQQQQPDSHQKIRRFYR----GDVIAIPAGTP 196
>BG646353 weakly similar to GP|12697782|db allergen Gly m Bd 28K {Glycine
max}, partial (24%)
Length = 696
Score = 34.3 bits (77), Expect = 0.098
Identities = 27/100 (27%), Positives = 37/100 (37%), Gaps = 2/100 (2%)
Frame = -3
Query: 341 LLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLS 400
L+ PH N RAT V G G ++V N + +
Sbjct: 694 LMTPHVNPRATEYGIVLRGSGRIQIVFPNGTNAMD---------------------THIK 578
Query: 401 PGDVYVIPAGHPIVVTASSD--LSLLGFGINAENNQRNFL 438
GDV+ IP ASS+ L GF +A+ N+ FL
Sbjct: 577 QGDVFFIPRYFAFCQIASSNEPLDFFGFTTSAQKNKPQFL 458
>TC84505 similar to PIR|T48442|T48442 hypothetical protein T32M21.60 -
Arabidopsis thaliana, partial (16%)
Length = 1130
Score = 33.5 bits (75), Expect = 0.17
Identities = 25/90 (27%), Positives = 42/90 (45%), Gaps = 8/90 (8%)
Frame = +2
Query: 234 IERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSRE-----QIEELSKNAK---SS 285
+E ++ ++ P DE R ++N ++ +E QI L +A+ +
Sbjct: 653 LENIVRGQSSTNPDATSNSNTDETRHDENQSNNLIDAQQENYDQEQIRSLETDARQFPNQ 832
Query: 286 SRRSESSESEPINLRNQKPIYSNKFGNFFE 315
+ SESS SEPIN + SN+ GN+ E
Sbjct: 833 TGTSESSTSEPINWQEA----SNQGGNWQE 910
>TC85216 similar to PIR|S26688|S26688 legumin K - garden pea, partial (66%)
Length = 1582
Score = 32.7 bits (73), Expect = 0.29
Identities = 30/131 (22%), Positives = 56/131 (41%), Gaps = 4/131 (3%)
Frame = +2
Query: 240 EENEQEPQHRRGLRKDERRQQSQEANV----IVKVSREQIEELSKNAKSSSRRSESSESE 295
EE E++P+ R G ++ER ++ K SRE+ +N + S + E
Sbjct: 659 EERERDPR-RPGHSQEEREREDDPYGRGRPWWEKESREKQRTRGQNGLEETICS-ARLVE 832
Query: 296 PINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVA 355
I +Y+ + G ++ P L++L + Y + + PH+N A ++
Sbjct: 833 NIARPAHADLYNPRAGRISDVNSLTLPILRNLRLSAEYVLLYRNGIYAPHWNINANSLLY 1012
Query: 356 VEEGKGEFELV 366
V G+G +V
Sbjct: 1013VIRGQGRVRIV 1045
>TC77076 similar to PIR|T07734|T07734 homeotic protein VAHOX1 - tomato,
partial (43%)
Length = 1664
Score = 31.6 bits (70), Expect = 0.64
Identities = 33/153 (21%), Positives = 65/153 (41%), Gaps = 5/153 (3%)
Frame = +1
Query: 234 IERVLIEENEQEPQHRRGLRKDERRQQSQEANVIV-KVSREQIEELSKNAKSSSRRSESS 292
+E+ E+N+ EP+ + L KD Q Q A + +R + ++L K+ S + ES
Sbjct: 676 LEKSFEEDNKLEPERKTKLAKDLGLQPRQVAIWFQNRRARWKTKQLEKDYDSLNDGYESL 855
Query: 293 ESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATV 352
++E NL +K ++ + E E+ Q + ++G F T+
Sbjct: 856 KTEYDNLLKEKDRLQSEVASLTEKVLEREKQ---------EGKFKQGESETKEFLKEPTI 1008
Query: 353 ----IVAVEEGKGEFELVGQRNENQQEQREYEE 381
+ +V EG+G + + + N + E+
Sbjct: 1009NKPLVDSVSEGEGSKLSIVEASNNNNNNNKLED 1107
>TC93604 weakly similar to SP|Q9SR72|GL32_ARATH Germin-like protein
subfamily 3 member 2 precursor. [Mouse-ear cress]
{Arabidopsis thaliana}, partial (28%)
Length = 500
Score = 31.6 bits (70), Expect = 0.64
Identities = 26/95 (27%), Positives = 41/95 (42%), Gaps = 2/95 (2%)
Frame = +3
Query: 279 SKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKN--PQLKDLDILVNYAEI 336
S++ K S + S ++ E K + K F + N P L L + A+
Sbjct: 174 SEDNKFSCKNSSTATVEDFTFSGIKLPGNFKDTGFSSMGVNSNVFPGLNTLGVSFVRADF 353
Query: 337 REGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNE 371
G + +PHF+ RAT + V EGK V +N+
Sbjct: 354 DAGGVNVPHFHPRATEVAIVLEGKIYSGFVDTKNK 458
>TC89462 similar to GP|9294563|dbj|BAB02826.1 gene_id:MQC12.22~unknown
protein {Arabidopsis thaliana}, partial (39%)
Length = 725
Score = 31.2 bits (69), Expect = 0.83
Identities = 24/87 (27%), Positives = 40/87 (45%), Gaps = 7/87 (8%)
Frame = +1
Query: 220 KNILEAAFNSNYEEIERVLIEENE----QEPQHRRGLRKDERRQQSQEANVIVKVSREQI 275
+NIL+A +++E V +EE E +E HR G+ + EA ++
Sbjct: 25 ENILDAIIYEE-DDVEMVDVEEGELVEEEEDHHRNGVAATQDHNLIHEAENNRNDINQKP 201
Query: 276 EELSKNAKSSSRRSESS---ESEPINL 299
+ LSKN K ++R + PIN+
Sbjct: 202 KALSKNRKRKNKRKRKALGGSDAPINI 282
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.133 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,309,366
Number of Sequences: 36976
Number of extensions: 142875
Number of successful extensions: 1089
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 1027
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1059
length of query: 440
length of database: 9,014,727
effective HSP length: 99
effective length of query: 341
effective length of database: 5,354,103
effective search space: 1825749123
effective search space used: 1825749123
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148289.2


