

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148289.14 - phase: 0
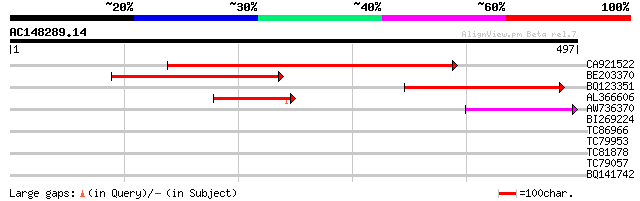
(497 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CA921522 289 2e-78
BE203370 277 8e-75
BQ123351 103 2e-22
AL366606 81 8e-16
AW736370 51 1e-06
BI269224 weakly similar to GP|11862972|db hypothetical protein {... 30 2.1
TC86966 similar to GP|6850934|emb|CAB71030.1 lectin-like protein... 29 4.8
TC79953 similar to GP|20279471|gb|AAM18751.1 unknown protein {Or... 29 4.8
TC81878 similar to GP|13877551|gb|AAK43853.1 Unknown protein {Ar... 28 6.2
TC79057 similar to GP|14581677|gb|AAK64512.1 Hsp70 interacting p... 28 8.1
BQ141742 weakly similar to PIR|T10798|T107 pherophorin-S - Volvo... 28 8.1
>CA921522
Length = 767
Score = 289 bits (739), Expect = 2e-78
Identities = 137/254 (53%), Positives = 178/254 (69%)
Frame = +3
Query: 139 KTSLIKEKLTLKGNLPSLPTKFLYQQASEFSKTNNVEAFYSILALLIYGLVLFPNIDNYV 198
+T +K L +G T FLY++ + F K AF SILALL+YGLVLFP++DN+V
Sbjct: 3 ETDEVKTHLITRGKFLGFSTDFLYERTTFFDKMGVAYAFNSILALLVYGLVLFPSLDNFV 182
Query: 199 DIHAIQIFLTKNPVPTLLADIYHSIHDRTQVGRGAILGCAPLLYKWFTSHLPQTHSFQAN 258
DI AIQIFL++NPVPTLL D Y SIH RTQ GRG IL CA LLY+W TSHLP+T F N
Sbjct: 183 DIKAIQIFLSRNPVPTLLGDTYLSIHRRTQAGRGTILCCAQLLYRWITSHLPRTPRFTTN 362
Query: 259 PENLSWPKRIMSLTPSDITWYRATCNFTNIIVSCGEYSNVPLLGTQGGISYNPILAKRQF 318
PENL W KR+MSLTP+++ WY + II SCG+++NVPLLG +GGISYNP LA+ QF
Sbjct: 363 PENLLWSKRLMSLTPAEVVWYDRVYDKGTIIDSCGKFANVPLLGMEGGISYNPTLARHQF 542
Query: 319 GCPMEAKPDNIYLQGEFYFNHEDPSNKRGRFVQAWHAIRTLNRSQLARRLDSLQGSYTQW 378
G PME KP +IYL+ +Y N +D + R V+AWH IR ++ QL ++ ++ SYTQW
Sbjct: 543 GYPMERKPLSIYLENVYYLNADDSTGMREHVVRAWHTIRRRDKDQLGKKTGAISSSYTQW 722
Query: 379 VVNRASDLVLPYHL 392
V++RA + +PY +
Sbjct: 723 VIDRAVQIGMPYKI 764
>BE203370
Length = 454
Score = 277 bits (708), Expect = 8e-75
Identities = 132/151 (87%), Positives = 144/151 (94%)
Frame = +1
Query: 90 HCFTFPDYQLMPTLEEYSHLIGLPVLDKVPFTGLEPFPKAATIANALHLKTSLIKEKLTL 149
HCFTFPDYQL PTLEEYS+L+G PVLDKVPFTG EP PKAATIA+ALHL+TSLIK KLT+
Sbjct: 1 HCFTFPDYQLAPTLEEYSYLVG*PVLDKVPFTGFEPIPKAATIADALHLETSLIKAKLTI 180
Query: 150 KGNLPSLPTKFLYQQASEFSKTNNVEAFYSILALLIYGLVLFPNIDNYVDIHAIQIFLTK 209
KGNLPSLPTKFLYQQAS+F+K +NV+AFYSILALLIYGLVLFPN+DNYVDIHAIQ+FLTK
Sbjct: 181 KGNLPSLPTKFLYQQASDFAKVDNVDAFYSILALLIYGLVLFPNVDNYVDIHAIQMFLTK 360
Query: 210 NPVPTLLADIYHSIHDRTQVGRGAILGCAPL 240
NPVPTLLAD+YHSIHDR QVGRGAILGCAPL
Sbjct: 361 NPVPTLLADMYHSIHDRNQVGRGAILGCAPL 453
>BQ123351
Length = 725
Score = 103 bits (256), Expect = 2e-22
Identities = 50/140 (35%), Positives = 88/140 (62%)
Frame = +2
Query: 347 GRFVQAWHAIRTLNRSQLARRLDSLQGSYTQWVVNRASDLVLPYHLPRYLSSTTPAPALP 406
G+F+QAW A+ + QL RR + SYT+WV++RA+ +P+ L R SSTT +P+LP
Sbjct: 2 GKFIQAWRAVHKREKGQLGRRS*LVYESYTKWVIDRAAKNGIPFPLQRLPSSTTLSPSLP 181
Query: 407 LAPATVEECQEKLARSECVSATWKRKYDEAMLKVETISGKIEQQEHEVHKLRRQIVKKNV 466
+ T EE Q LA TW+ +Y EA ++ T+ G++EQ++HE+ K+R+Q+++++
Sbjct: 182 MTSKTKEEAQYLLAEMTREKDTWRIRYMEAESEIGTLRGQVEQKDHELLKMRQQMIERDD 361
Query: 467 QIRAQSTRLSQFISAGERWE 486
++ + L + I+ +R +
Sbjct: 362 LLQEKDKLLKKHITKKQRMD 421
>AL366606
Length = 393
Score = 81.3 bits (199), Expect = 8e-16
Identities = 42/78 (53%), Positives = 54/78 (68%), Gaps = 6/78 (7%)
Frame = -1
Query: 179 SILALLIYGLVLFPNIDNYVDIHAIQIFLTKNPVPTLLADIYHSIHDRTQVGRGAILGCA 238
+I L GL+LFPN+DN+VD++AI++F +KNPVPTLLAD YH+IHDRT GRG IL
Sbjct: 381 AIXLCLSTGLILFPNLDNFVDMNAIEVFHSKNPVPTLLADTYHAIHDRTLKGRGYILWLH 202
Query: 239 PLL------YKWFTSHLP 250
L ++ F S LP
Sbjct: 201 ILFSNRVVYFRTFRSSLP 148
>AW736370
Length = 488
Score = 50.8 bits (120), Expect = 1e-06
Identities = 31/99 (31%), Positives = 49/99 (49%), Gaps = 1/99 (1%)
Frame = -2
Query: 400 TPAPALPLAPATVEECQEKLARSECVSATWKRKYDEAMLKVETISGKIEQQEHEVHKLRR 459
TPAP LP+ T +E QE+L +E WKR+Y + ET+ G +EQ+ ++
Sbjct: 481 TPAPPLPMTFDTEKEYQERLTEAEREIHRWKREYQKKDQDYETVMGLLEQEAYDSR*KDV 302
Query: 460 QIVKKNVQIRAQSTRLSQFISAGE-RWEFFKDAHSDSDE 497
I K N +I+ L + + R + F HSD ++
Sbjct: 301 IIAKLNERIKENDAALDRIPGRKKRRMDLFDGPHSDFED 185
>BI269224 weakly similar to GP|11862972|db hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (26%)
Length = 537
Score = 30.0 bits (66), Expect = 2.1
Identities = 16/49 (32%), Positives = 27/49 (54%)
Frame = -3
Query: 432 KYDEAMLKVETISGKIEQQEHEVHKLRRQIVKKNVQIRAQSTRLSQFIS 480
KY E M + S + + +V+ ++VKKN ++ QST+ S+ IS
Sbjct: 490 KYTEVMTSR*SCSRQYITNDDDVNNKNTKVVKKNRNLKKQSTKTSKSIS 344
>TC86966 similar to GP|6850934|emb|CAB71030.1 lectin-like protein {Cicer
arietinum}, partial (82%)
Length = 942
Score = 28.9 bits (63), Expect = 4.8
Identities = 19/73 (26%), Positives = 32/73 (43%)
Frame = -3
Query: 184 LIYGLVLFPNIDNYVDIHAIQIFLTKNPVPTLLADIYHSIHDRTQVGRGAILGCAPLLYK 243
LI+ LFPN ++ ++ +P T+L + +H H C P LY
Sbjct: 385 LIFI*SLFPNFSHHK-------YICASPTETILLNFHHKFH----------YVCIPFLYF 257
Query: 244 WFTSHLPQTHSFQ 256
+FT +L + +Q
Sbjct: 256 YFTRYLQP*NLYQ 218
>TC79953 similar to GP|20279471|gb|AAM18751.1 unknown protein {Oryza sativa
(japonica cultivar-group)}, partial (25%)
Length = 899
Score = 28.9 bits (63), Expect = 4.8
Identities = 12/37 (32%), Positives = 23/37 (61%)
Frame = -3
Query: 449 QQEHEVHKLRRQIVKKNVQIRAQSTRLSQFISAGERW 485
+Q H RR+IV + + IR S+R+++ +++ RW
Sbjct: 498 RQRHRNSSRRRRIVIRIIMIRRSSSRVNKSVTSSLRW 388
>TC81878 similar to GP|13877551|gb|AAK43853.1 Unknown protein {Arabidopsis
thaliana}, partial (62%)
Length = 911
Score = 28.5 bits (62), Expect = 6.2
Identities = 22/91 (24%), Positives = 42/91 (45%), Gaps = 8/91 (8%)
Frame = +2
Query: 3 SFIILSFLIKNIEGLILVFVNII--------GMERRNTRKFTFRKLDLENLKKLAFEVTS 54
+F+IL L+ + GL+LVF+ ++ G+ ++ F KL L+ ++ L
Sbjct: 275 TFMILPQLMVTLIGLVLVFIILVFKFMVLNMGLVHMIVKRRGFLKLSLDTVQDLPLGNQF 454
Query: 55 LENFRDRHGNLLGVLWTNIEKGCLETLVQFY 85
+ ++ + G LW + K LE + Y
Sbjct: 455 TLDQQNWDRRMFGNLWRS*LKSMLEIHIILY 547
>TC79057 similar to GP|14581677|gb|AAK64512.1 Hsp70 interacting
protein/thioredoxin chimera {Vitis labrusca}, partial
(84%)
Length = 1335
Score = 28.1 bits (61), Expect = 8.1
Identities = 18/52 (34%), Positives = 27/52 (51%), Gaps = 4/52 (7%)
Frame = +1
Query: 430 KRKYDE----AMLKVETISGKIEQQEHEVHKLRRQIVKKNVQIRAQSTRLSQ 477
K YDE A+ KVE + KIE+ + +LR+Q +K Q + Q +Q
Sbjct: 622 KIDYDEEIAMALKKVEPNAHKIEEHRKKYERLRKQKEQKRAQPKKQPQNQAQ 777
>BQ141742 weakly similar to PIR|T10798|T107 pherophorin-S - Volvox carteri,
partial (9%)
Length = 1337
Score = 28.1 bits (61), Expect = 8.1
Identities = 16/44 (36%), Positives = 19/44 (42%)
Frame = -3
Query: 113 PVLDKVPFTGLEPFPKAATIANALHLKTSLIKEKLTLKGNLPSL 156
PVL + PFP +A I A HL+ L LPSL
Sbjct: 903 PVLSRSLILTYPPFPASAVILRASHLRYRFAPSYLRSSAPLPSL 772
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.137 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,729,691
Number of Sequences: 36976
Number of extensions: 259366
Number of successful extensions: 1261
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 1249
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1260
length of query: 497
length of database: 9,014,727
effective HSP length: 100
effective length of query: 397
effective length of database: 5,317,127
effective search space: 2110899419
effective search space used: 2110899419
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148289.14


