

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148289.13 - phase: 0
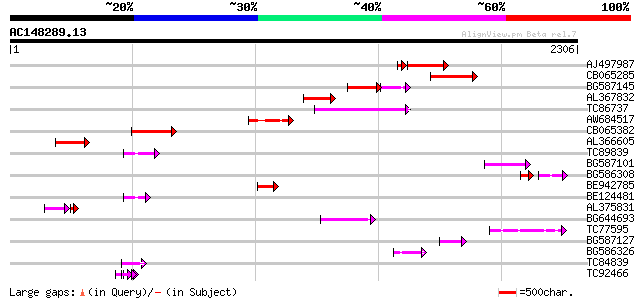
(2306 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 351 2e-96
CB065285 weakly similar to PIR|A84500|A84 probable retroelement ... 302 1e-81
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 157 2e-58
AL367832 weakly similar to GP|14091845|gb Putative retroelement ... 196 9e-50
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 179 1e-44
AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thalia... 174 4e-43
CB065382 similar to PIR|T09671|T096 RPE15 protein - alfalfa (fra... 162 1e-39
AL366605 136 8e-32
TC89839 homologue to SP|P23466|CYAA_SACKL Adenylate cyclase (EC ... 114 3e-25
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 109 1e-23
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 66 3e-19
BE942785 92 2e-18
BE124481 92 2e-18
AL375831 74 9e-18
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 84 6e-16
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 77 1e-13
BG587127 weakly similar to PIR|H84506|H84 probable retroelement ... 68 5e-11
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 53 2e-06
TC84839 similar to GP|19386827|dbj|BAB86205. hypothetical protei... 53 2e-06
TC92466 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis... 51 6e-06
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 351 bits (901), Expect = 2e-96
Identities = 166/167 (99%), Positives = 166/167 (99%)
Frame = -2
Query: 1617 KTCCALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPALTGRIARWQMLLSEYDIECRS 1676
KTCCALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPALTGRIARWQMLLSEYDIE RS
Sbjct: 635 KTCCALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPALTGRIARWQMLLSEYDIEYRS 456
Query: 1677 QKAIKGSILADHLAHQPLEDYRPIKFDFPDEEIMYLKMKDCDEPLFGEGPDPDSVWGLIF 1736
QKAIKGSILADHLAHQPLEDYRPIKFDFPDEEIMYLKMKDCDEPLFGEGPDPDSVWGLIF
Sbjct: 455 QKAIKGSILADHLAHQPLEDYRPIKFDFPDEEIMYLKMKDCDEPLFGEGPDPDSVWGLIF 276
Query: 1737 DGAVNVYGNGIGAVLLTPKGAHIPFTARLRFDCTNNIAEYEACIMGI 1783
DGAVNVYGNGIGAVLLTPKGAHIPFTARLRFDCTNNIAEYEACIMGI
Sbjct: 275 DGAVNVYGNGIGAVLLTPKGAHIPFTARLRFDCTNNIAEYEACIMGI 135
Score = 50.4 bits (119), Expect = 8e-06
Identities = 24/35 (68%), Positives = 26/35 (73%)
Frame = +2
Query: 1579 NSMGCVLGQQDETGRKEHAIYYLSKKFTECESRYS 1613
N CVLGQQDETGRKEHAIYYLS+K + R S
Sbjct: 26 NPWVCVLGQQDETGRKEHAIYYLSRKLSSTVWRIS 130
>CB065285 weakly similar to PIR|A84500|A84 probable retroelement gag/pol
polyprotein [imported] - Arabidopsis thaliana, partial
(2%)
Length = 592
Score = 302 bits (773), Expect = 1e-81
Identities = 143/196 (72%), Positives = 168/196 (84%), Gaps = 8/196 (4%)
Frame = -2
Query: 1713 KMKDCDEPLFGEGPDPDSVWGLIFDGAVNVYGNGIGAVLLTPKGAHIPFTARLRFDCTNN 1772
K KDC+EPL GEGPDP+S WGL+FDGAVN YG GIGAV+++P+G +IPFTAR+ F+CTNN
Sbjct: 591 KSKDCEEPLIGEGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGHYIPFTARILFECTNN 412
Query: 1773 IAEYEACIMGIEEAIDLRIKNIEIYGDSALVINQIKGKWETLHAGLIPYRDYARRLLTFF 1832
+AEYEACI GIEEAID+RIK+++IYGDSALVINQIKG+WET HA LIPYRDYARRLLT+F
Sbjct: 411 MAEYEACIFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHANLIPYRDYARRLLTYF 232
Query: 1833 NKVELHHIPRDENQMADALATLSSMIKVNHHNDVPLISVKFLDRPAYVFA--------AE 1884
KVELHHIPRDENQMADALATLSSM +VNH NDVP+I V+ L+RP++VFA E
Sbjct: 231 TKVELHHIPRDENQMADALATLSSMFRVNHWNDVPIIKVQRLERPSHVFAIGNVIDQTGE 52
Query: 1885 VVFDDKPWFHDIKVFL 1900
V D KPW++DIK FL
Sbjct: 51 NVVDYKPWYYDIKQFL 4
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 157 bits (397), Expect(2) = 2e-58
Identities = 72/136 (52%), Positives = 104/136 (75%)
Frame = +2
Query: 1374 MAPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEIEVYVDDMIVK 1433
M P+D EKT+FIT GT+CYKVMPFGL NAG+TYQR + +F D + +EVY+DDM+VK
Sbjct: 2 MHPDDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVK 181
Query: 1434 SVTEEDHVKYLQKMFQRLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVKAIR 1493
S+ DH+ +L++ F+ L +Y ++LNP KCTFGV SG+ LG+IV+Q+GIEV+P ++ AI
Sbjct: 182 SLRATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAIL 361
Query: 1494 EMPAPRTEKEVRGFLG 1509
++P+P+ +EV+ G
Sbjct: 362 DLPSPKNSREVQRLTG 409
Score = 89.4 bits (220), Expect(2) = 2e-58
Identities = 45/120 (37%), Positives = 72/120 (59%)
Frame = +3
Query: 1509 GRLNYISRFISHMTATCGPIFKLLRKEQGIVWTEDCQKAFDSIKKYLLEPPILIPPVEGR 1568
GR+ ++RFIS T C P +KLL + VW E C++AF+ +K+YL PP+L P G
Sbjct: 408 GRIAALNRFISRSTDKCLPFYKLLCGNKRFVWDEKCEEAFEQLKQYLTTPPVLSKPEAGD 587
Query: 1569 PLIMYLTVLENSMGCVLGQQDETGRKEHAIYYLSKKFTECESRYSMLEKTCCALAWAAKR 1628
L +Y+ + ++ VL ++D +K I+Y SK+ T+ E+RY LEK A+ +A++
Sbjct: 588 TLSLYIAISSTAVSSVLIREDRGEQK--PIFYTSKRMTDPETRYPTLEKMAFAVITSARK 761
>AL367832 weakly similar to GP|14091845|gb Putative retroelement {Oryza
sativa}, partial (2%)
Length = 384
Score = 196 bits (498), Expect = 9e-50
Identities = 90/128 (70%), Positives = 112/128 (87%)
Frame = -1
Query: 1195 QEKKTIQPYGDELEVINLGTKEDKKEIKVGASLETSVKKQVIELLKEYVDVFAWSYQDMP 1254
QE+K IQP+ +E+E+INLGT+E+K+EIKVGA+LE VK+++ +LL+EY+D+FA SY+DMP
Sbjct: 384 QERKAIQPHQEEIELINLGTEENKREIKVGAALEEGVKRKIFQLLREYLDIFACSYEDMP 205
Query: 1255 GLDTDIVVHHLPLKPECPPVKQKLRRTRPDMALKIKEEVQKQIDAGFLITSNYPQWLANI 1314
GLD IV H +P KPECPPV+ KLRRT PDMALKIK EVQKQIDAGFL+T YP+W+ANI
Sbjct: 204 GLDPKIVEHRIPTKPECPPVR*KLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANI 25
Query: 1315 VPVPKKDG 1322
VPVPKKDG
Sbjct: 24 VPVPKKDG 1
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 179 bits (453), Expect = 1e-44
Identities = 121/398 (30%), Positives = 205/398 (51%), Gaps = 7/398 (1%)
Frame = +1
Query: 1238 LLKEYVDVFAWSYQDMPGLDTDIVVHHLPLKPEC----PPVKQ-KLRRTRPDMALKIKEE 1292
+L+E+ D+F ++ H +PL P+ PP+ L L +K+
Sbjct: 343 VLEEFPDLFNPEKAYQVPASRGLLDHAIPLIPDKDGNDPPLPWGPLYGMSRQELLVLKKT 522
Query: 1293 VQKQIDAGFLITSNYPQWLANIVPVPKKDGKVRMCVDYRDLNKASPKDDFPLPHIDVLVD 1352
++ +D GF+ S A ++ V K G +R CVDYR LN + KD +PLP I +
Sbjct: 523 LEDLLDKGFIKASGSAAG-APVLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISETLR 699
Query: 1353 STAKSKVFSFMDGFSGYNQIKMAPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMT 1412
A ++ F+ +D + ++++++ ED+EKT+F T +G F + V PFGL A AT+QR +
Sbjct: 700 RVAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQRYIN 879
Query: 1413 TLFHDMIHKEIEVYVDDMIVKSV-TEEDHVKYLQKMFQRLRKYKLRLNPNKCTFGVRSGK 1471
H+ + + Y+DD+++ + +++DH ++++ +RL L L+P KC F V + K
Sbjct: 880 KTLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVTTVK 1059
Query: 1472 LLGFIVSQ-KGIEVDPDKVKAIREMPAPRTEKEVRGFLGRLNYISRFISHMTATCGPIFK 1530
+GFI++ KG+ DP K+ AIR+ P + K R FLG NY FI + P+ +
Sbjct: 1060 YVGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGARSFLGFCNYYKDFIPGYSEITEPLTR 1239
Query: 1531 LLRKEQGIVWTEDCQKAFDSIKKYLLEPPILIPPVEGRPLIMYLTVLENSMGCVLGQQDE 1590
L RK+ W + + AF +K+ E P+L + ++G VL Q+D
Sbjct: 1240 LTRKDFPFRWGAEQEAAFTKLKRLFAEEPVLRMFDPEAVTTVETDCSGFALGGVLTQEDG 1419
Query: 1591 TGRKEHAIYYLSKKFTECESRYSMLEKTCCALAWAAKR 1628
TG H + + S++ + E Y + +K A+ WA R
Sbjct: 1420 TG-AAHPVAFHSQRLSPAEYNYPIHDKELLAV-WACLR 1527
>AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thaliana chromosome
II BAC F26H6; putative retroelement pol polyprotein,
partial (1%)
Length = 488
Score = 174 bits (441), Expect = 4e-43
Identities = 92/183 (50%), Positives = 119/183 (64%), Gaps = 1/183 (0%)
Frame = +1
Query: 970 VNFQVMDIQASYSCLLGRPWIHEAGAVTSTLHQKLKFVKNGKLVTVNGEEALLVSHLSSF 1029
+ FQVMDI ASYSCLLGRPWIH+AGAVTSTLHQKLKFVKNG
Sbjct: 1 ITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKFVKNG------------------- 123
Query: 1030 SFIGADNVEGTPFQGFTIEDKNTKRNEASISSLKDAQKVIQAGGSTSWGKLIELPENKHR 1089
+ EGT FQG ++E K+ A+++SLKDAQK +Q G + WGKLI+L ENK +
Sbjct: 124 ------SAEGTAFQGLSMEGAEPKKVGAAMASLKDAQKAVQEGQAADWGKLIQLCENKRK 285
Query: 1090 EGLGFFPSTGLSTAKKGTFHSSGFIHAIIGDDPESVPRG-FITPGVSSHNWVAIDVPFVA 1148
EGL F P++G+ST GTFHS+GF++ + + VPR F+ PG + +W A+DVP +
Sbjct: 286 EGLRFSPTSGVST---GTFHSAGFVNTLAEEVARFVPRPLFVIPGGIAKDWDAVDVPSIM 456
Query: 1149 HLS 1151
H+S
Sbjct: 457 HVS 465
>CB065382 similar to PIR|T09671|T096 RPE15 protein - alfalfa (fragment),
partial (9%)
Length = 624
Score = 162 bits (410), Expect = 1e-39
Identities = 87/182 (47%), Positives = 113/182 (61%)
Frame = -2
Query: 495 PQQNFQQQPYQQRPQQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPPIPEKLPSWY 554
P +N +QQR Q P P+ IP+ YAE LP LL + R P P+ LP +
Sbjct: 539 PPRNNHPPVHQQRQ*QQARPTFPL--IPMLYAE*LPTLLLRGHCTIRQGKPPPDPLPPRF 366
Query: 555 RLDQTCDFHEGGRGHNIETCYAFKSTVQKLINDGKITFTDSAPNVQTNPLPNHGAATVNM 614
R D CDFH+G GH++E CYA K V+KLIN GK+TF ++ P+V NPLPNH A VNM
Sbjct: 365 RSDLKCDFHQGALGHDVEGCYALKHIVKKLINQGKLTFENNVPHVLDNPLPNH--AAVNM 192
Query: 615 IEDCQKTRPILNVQRIRTPLVPLHAKLCKVDLFEHDHDLCEICLMNSGGCQKVRNDIQGL 674
IE ++ P L+V+ + TPLVPLH KLC+ LF+HDH C+ N GC V+NDIQ L
Sbjct: 191 IEVYEEA-PGLDVRNVTTPLVPLHIKLCQASLFDHDHANCQE*FYNPLGCCVVQNDIQSL 15
Query: 675 LD 676
++
Sbjct: 14 MN 9
>AL366605
Length = 422
Score = 136 bits (343), Expect = 8e-32
Identities = 65/140 (46%), Positives = 93/140 (66%)
Frame = -3
Query: 186 DMKALREKGKFGKTAYDLCLVPSVQVPHKFKIPDFEKYKGSSCPEEHLKMYVRRMPAYAQ 245
++KA R KT DLCLV V VP KFKIPDF++Y G +CP+ H+ YVR+M Y
Sbjct: 417 EIKANRGNADSFKTQ-DLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYKD 241
Query: 246 DDQILIYYFQESLTGPASKWYTNLDKTRIQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMT 305
+D ++I+ FQ+SL A++WYT+L K I TF +L AF Y +N + P+R L++++
Sbjct: 240 NDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDELAAAFKSHYGFNTRLKPNREFLRSLS 61
Query: 306 QGDKETFKEYAQRWRDTAAQ 325
Q +E+F+EYAQRWR AA+
Sbjct: 60 QKKEESFREYAQRWRGAAAR 1
>TC89839 homologue to SP|P23466|CYAA_SACKL Adenylate cyclase (EC 4.6.1.1)
(ATP pyrophosphate-lyase) (Adenylyl cyclase). [Yeast],
partial (0%)
Length = 665
Score = 114 bits (286), Expect = 3e-25
Identities = 69/155 (44%), Positives = 83/155 (53%), Gaps = 7/155 (4%)
Frame = +3
Query: 463 PQSVQQQNFQQQPYQ----QQPYQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPRMPI 518
P VQQ+ Q PYQ Q+P QQ P Q P Q PQ+ Q Q+ Q
Sbjct: 228 PPLVQQR--PQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQ--------C 377
Query: 519 NPIPVTYAELLPGLLKKNLVQTRTAPPIPEKLPSWYRLDQTCDFHEGGRGHNIETCYAFK 578
+PIPV YA+LLP LLKKNL+QT P +P LP WYR D C FH+G GH+ E CY K
Sbjct: 378 DPIPVKYADLLPILLKKNLIQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLK 557
Query: 579 STVQKLINDGKITFTDSAPNV---QTNPLPNHGAA 610
VQKLI + +F D V Q + P+ GAA
Sbjct: 558 EEVQKLIENNVWSFDDQDIKVLLQQQHLAPHSGAA 662
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 109 bits (273), Expect = 1e-23
Identities = 60/188 (31%), Positives = 95/188 (49%)
Frame = +2
Query: 1931 LYKRNFDTVLLRCVDKYEADLLIHEIHEGSFGIHPNGHTMAKKILRAGYYWMTMESDCYK 1990
LYK+ D + +RCV + E ++ H ++ H KI +AG++W TM D +
Sbjct: 41 LYKQCADNIYIRCVAEEEIPGILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHS 220
Query: 1991 HTRKCHKCQIYADKIHMPPTTLNLLSSPWPFSMWGIDMIGRIEPKASNGHRFILVAIDYF 2050
KC CQ + N + F +WGID +G +N ++ILVA+DY
Sbjct: 221 FISKCDPCQRQGNIS*RNEMPQNFILEVEVFDVWGIDFMGPFPSSYNN--KYILVAVDYV 394
Query: 2051 TKWVEAASYANVTKQVVVKFIKNHIICRYGIPNRIITDNGTNLNNKMMKELCDDFKIEHH 2110
+KWVEA + VVVK K+ I R+G+P +I+D G++ NK+ ++L + H
Sbjct: 395 SKWVEAIASPTNDATVVVKMFKSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHK 574
Query: 2111 NSSPYRPQ 2118
++ Y PQ
Sbjct: 575 VATAYHPQ 598
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 686
Score = 65.9 bits (159), Expect(2) = 3e-19
Identities = 42/122 (34%), Positives = 69/122 (56%), Gaps = 4/122 (3%)
Frame = -3
Query: 2152 LHGYRTSVRTSTGATPFSLVYGMEAVLPVEVEIPSL---RVLMEADLSEAEWVQNRYDQL 2208
L +RT+ R +T +TPFS+ + +EA+ P EV + SL R+ +L+ ++ L
Sbjct: 462 LWSHRTNPRGATKSTPFSMAHRVEAMAPAEVNVTSL*RSRMPQNIELNN----DRLFNAL 295
Query: 2209 NLIEEKRMTALCHGQLYQKRMKQAFDKKVRPREFKEGDLVLKKIFSFQPD-SRGKWAPNY 2267
IEE+R AL Q YQ +++ ++K V+ + K GD+VL K+F + + GK N+
Sbjct: 294 ETIEERRDQALLRIQNYQHQIESYYNKTVKSQPLKLGDIVLCKVFENTKELNAGKLGTNW 115
Query: 2268 EG 2269
EG
Sbjct: 114 EG 109
Score = 49.7 bits (117), Expect(2) = 3e-19
Identities = 21/52 (40%), Positives = 34/52 (65%)
Frame = -2
Query: 2079 YGIPNRIITDNGTNLNNKMMKELCDDFKIEHHNSSPYRPQMNGAVEAANKNI 2130
+G+P I+TDNG++ + +E C+ ++I + +SP PQ NG EA+NK I
Sbjct: 685 HGLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKII 530
>BE942785
Length = 460
Score = 92.4 bits (228), Expect = 2e-18
Identities = 43/82 (52%), Positives = 60/82 (72%)
Frame = -2
Query: 1009 NGKLVTVNGEEALLVSHLSSFSFIGADNVEGTPFQGFTIEDKNTKRNEASISSLKDAQKV 1068
+G LVT++GEEA L+S LSSFS I A + EGT FQG T+E KR+ +++SLKDAQ+
Sbjct: 378 SGXLVTIHGEEAYLISQLSSFSCIEAGSAEGTAFQGLTVEGTEPKRDGTAMASLKDAQRA 199
Query: 1069 IQAGGSTSWGKLIELPENKHRE 1090
+Q + WG+LI+L ENKH++
Sbjct: 198 VQESQAAGWGRLIQLRENKHKD 133
>BE124481
Length = 538
Score = 92.0 bits (227), Expect = 2e-18
Identities = 52/112 (46%), Positives = 62/112 (54%), Gaps = 4/112 (3%)
Frame = +3
Query: 463 PQSVQQQNFQQQPYQ----QQPYQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPRMPI 518
P VQQ+ Q PYQ Q+P QQ P Q P Q PQ+ Q Q+ Q
Sbjct: 231 PPLVQQR--PQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQ--------C 380
Query: 519 NPIPVTYAELLPGLLKKNLVQTRTAPPIPEKLPSWYRLDQTCDFHEGGRGHN 570
+PIPV YA+LLP LLKKNL+QT P +P LP WYR D C FH+G GH+
Sbjct: 381 DPIPVKYADLLPILLKKNLIQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHD 536
>AL375831
Length = 467
Score = 73.6 bits (179), Expect(2) = 9e-18
Identities = 39/102 (38%), Positives = 59/102 (57%), Gaps = 1/102 (0%)
Frame = +2
Query: 143 TMTYSAPVIHTVPQTEEPIFHSGNVEAYEEVNDLREKY-DELRRDMKALREKGKFGKTAY 201
T + P++HT+PQ ++ + + ++ E+ ELR +++A R KT
Sbjct: 56 TAAVTEPLVHTLPQGININTQHRSIPVTKTMEEMMEELAKELRHEIQANRGNADSVKTQ- 232
Query: 202 DLCLVPSVQVPHKFKIPDFEKYKGSSCPEEHLKMYVRRMPAY 243
DLCLV V VP KFKIPDF++Y G +CP+ H+ YVR+M Y
Sbjct: 233 DLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNY 358
Score = 37.0 bits (84), Expect(2) = 9e-18
Identities = 15/32 (46%), Positives = 23/32 (71%)
Frame = +3
Query: 246 DDQILIYYFQESLTGPASKWYTNLDKTRIQTF 277
+D ++I+ FQ+SL A++WYT+L K I TF
Sbjct: 366 NDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTF 461
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 84.0 bits (206), Expect = 6e-16
Identities = 64/226 (28%), Positives = 108/226 (47%), Gaps = 5/226 (2%)
Frame = +2
Query: 1265 LPLKPECPPVKQKLRRTRPDMALKIKEEVQKQIDAGFLITSNYPQWLANIVPVPKKDGKV 1324
+ L P P+ R P +K +++ ++ GF+ S YP + ++ + KKDG +
Sbjct: 47 IDLLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVV-VLFLKKKDGFL 223
Query: 1325 RMCVDYRDLNKASPKDDFPLPHIDVLVDSTAKSKVFSFMDGFSGYNQIKMAPEDREKTSF 1384
RM +DY LN + K +PLP ID L D+ SK F +D G +Q ++ ED KT+F
Sbjct: 224 RMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVIGEDVPKTAF 403
Query: 1385 ITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEIEVYVDDMIVKSVTEEDHVKYL 1444
+G + VM FG N + M +F D + + V+ +D+++ S E +H +L
Sbjct: 404 RIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSKNENEHENHL 583
Query: 1445 QKMFQRLRKYKLRLNPNKCTFG-----VRSGKLLGFIVSQKGIEVD 1485
+ + L+ L C V G ++S +G++VD
Sbjct: 584 RLALKVLKDIGL------CQISYV*ILVEVGFFSLHVISGEGLKVD 703
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 76.6 bits (187), Expect = 1e-13
Identities = 72/325 (22%), Positives = 134/325 (41%), Gaps = 13/325 (4%)
Frame = +2
Query: 1952 LIHEIHEGSFGIHPNGHTMAKKILRAGYYWMTMESDCYKHTRKCHKCQIYADKIHMPPTT 2011
L+ E H+ + HP G +I+ ++W + R C C IH+
Sbjct: 179 LVQESHDSTAAGHP-GRNGTLEIVSRKFFWPGQSQTVRRFVRNCDVC----GGIHIWRQA 343
Query: 2012 LNLLSSPWPF-----SMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYANVTKQV 2066
P P S +D I + P G +++ V +D +K V + +
Sbjct: 344 KRGFLKPLPVPNRLHSDLSMDFITSLPPTRGRGSQYLWVIVDRLSKSVTLEEMDTMEAEA 523
Query: 2067 VVKFIKNHIICRY---GIPNRIITDNGTNLNNKMMKELCDDFKIEHHNSSPYRPQMNGAV 2123
+ + C Y G+P I++D G+N + +E C + S+ Y PQ +G
Sbjct: 524 CAQ---RFLSCHYRFHGMPQSIVSDRGSNWVGRFWREFCRLTGVTQLLSTSYHPQTDGGT 694
Query: 2124 EAANKNIKRIVQKMVVTYKD-WHEMLPFALHGYRTSVRTSTGATPFSLVYGMEAVLPVEV 2182
E N+ I+ +++ V +D W ++LP R +S GATPF + +G V P+
Sbjct: 695 ERWNQEIQAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSIGATPFFVEHGYH-VDPIPT 871
Query: 2183 EIPSLRVLMEADLSEAEWVQNRYDQLNLIEEKRMTALCHGQLYQKRMKQAFDKKVRPRE- 2241
+ V+ E + + V+ D I+ + + A Q+R + + +K+ P +
Sbjct: 872 VEDTGGVVSEGEAAAQLLVKRMKDVTGFIQAEIVAA-------QQRSEASANKRRCPADR 1030
Query: 2242 FKEGDLVLKKIFSF---QPDSRGKW 2263
++ GD V + ++ +P + W
Sbjct: 1031 YQVGDKVWLNVSNYKSPRPSKKLDW 1105
>BG587127 weakly similar to PIR|H84506|H84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(13%)
Length = 415
Score = 67.8 bits (164), Expect = 5e-11
Identities = 41/109 (37%), Positives = 56/109 (50%)
Frame = +3
Query: 1748 GAVLLTPKGAHIPFTARLRFDCTNNIAEYEACIMGIEEAIDLRIKNIEIYGDSALVINQI 1807
G L +P + + RL F +NN YEA I G+ A L+I+NI Y DS LV +Q
Sbjct: 3 GIRLTSPTNEILKQSFRLEFHASNNETNYEALIAGVRLAHGLKIRNIHAYCDSQLVASQF 182
Query: 1808 KGKWETLHAGLIPYRDYARRLLTFFNKVELHHIPRDENQMADALATLSS 1856
G++E + Y ++L + L IPR EN ADALA L+S
Sbjct: 183 SGEYEARDELMDTYLKLVQKLAQKLDYFALTRIPRSENVQADALAALAS 329
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 52.8 bits (125), Expect = 2e-06
Identities = 37/137 (27%), Positives = 64/137 (46%)
Frame = +2
Query: 1559 PILIPPVEGRPLIMYLTVLENSMGCVLGQQDETGRKEHAIYYLSKKFTECESRYSMLEKT 1618
PIL+ P E ++Y +GCVL Q E I Y S++ + E Y +
Sbjct: 17 PILVLP-ELITYVVYTDASITGLGCVLTQH------EKVIAYASRQLRKHEGNYPTHDLE 175
Query: 1619 CCALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPALTGRIARWQMLLSEYDIECRSQK 1678
A+ +A K R Y+ + + +KYIF +P L R RW +++YD++
Sbjct: 176 MAAVVFALKIWRSYLYGAKVQIHTDHKSLKYIFTQPELNLRQRRWMEFVADYDLDITYYP 355
Query: 1679 AIKGSILADHLAHQPLE 1695
K +++AD L+ + ++
Sbjct: 356 G-KANLVADALSRRRVD 403
>TC84839 similar to GP|19386827|dbj|BAB86205. hypothetical protein~similar
to Arabidopsis thaliana chromosome 5 T6I14_10, partial
(20%)
Length = 819
Score = 52.8 bits (125), Expect = 2e-06
Identities = 42/108 (38%), Positives = 46/108 (41%), Gaps = 5/108 (4%)
Frame = +1
Query: 453 PQNPSPQNVQPQSVQQQNFQQQPYQQQPYQQQP----YQQYPYQQYPQQNFQQQPYQQRP 508
P NP Q Q Q QQQ QQQ YQQ QQQP +QQ Q P +F + P P
Sbjct: 163 PHNPQHQQHQ-QHQQQQQQQQQQYQQHQQQQQPPHFHHQQQQQQSGPGPDFHRGPPPPMP 339
Query: 509 QQPRP-PRMPINPIPVTYAELLPGLLKKNLVQTRTAPPIPEKLPSWYR 555
QQP P R P +E LPG PP P P YR
Sbjct: 340 QQPPPMMRQPSASSTNLGSEFLPG-----------GPPGPPGPPPHYR 450
>TC92466 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis thaliana},
partial (14%)
Length = 632
Score = 50.8 bits (120), Expect = 6e-06
Identities = 28/60 (46%), Positives = 33/60 (54%)
Frame = +1
Query: 454 QNPSPQNVQPQSVQQQNFQQQPYQQQPYQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRP 513
Q Q QPQ Q Q+ QQQ Q QQ Q++ QQ QQ QQQP Q +PQQP+P
Sbjct: 379 QQQQQQQQQPQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQP-QSQPQQPQP 555
Score = 45.8 bits (107), Expect = 2e-04
Identities = 28/66 (42%), Positives = 33/66 (49%)
Frame = +1
Query: 432 QQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQSVQQQNFQQQPYQQQPYQQQPYQQYPY 491
+Q Q Q PQ PQ Q Q Q++Q Q + Q QQ QQQ +QQQP Q P
Sbjct: 370 EQQQQQQQQQQQPQ-PQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQP-QSQPQ 543
Query: 492 QQYPQQ 497
Q PQQ
Sbjct: 544 QPQPQQ 561
Score = 45.8 bits (107), Expect = 2e-04
Identities = 26/61 (42%), Positives = 28/61 (45%)
Frame = +1
Query: 462 QPQSVQQQNFQQQPYQQQPYQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPRMPINPI 521
Q Q QQQ Q QP Q Q QQQ Q QQ Q QQ QQ+ Q +P P P
Sbjct: 373 QQQQQQQQQQQPQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQPQSQPQQPQ 552
Query: 522 P 522
P
Sbjct: 553 P 555
Score = 43.5 bits (101), Expect = 0.001
Identities = 27/69 (39%), Positives = 32/69 (46%)
Frame = +1
Query: 439 QIPQYPQIPQYPQFPQNPSPQNVQPQSVQQQNFQQQPYQQQPYQQQPYQQYPYQQYPQQN 498
Q Q Q Q PQ Q+ Q Q Q +Q Q Q + QQ QQQ +QQ P Q Q
Sbjct: 373 QQQQQQQQQQQPQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQPQSQPQQPQ 552
Query: 499 FQQQPYQQR 507
QQQ + R
Sbjct: 553 PQQQQNRDR 579
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 70,598,720
Number of Sequences: 36976
Number of extensions: 1063284
Number of successful extensions: 9821
Number of sequences better than 10.0: 170
Number of HSP's better than 10.0 without gapping: 6670
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8943
length of query: 2306
length of database: 9,014,727
effective HSP length: 112
effective length of query: 2194
effective length of database: 4,873,415
effective search space: 10692272510
effective search space used: 10692272510
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC148289.13


