

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148176.1 + phase: 0
(700 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
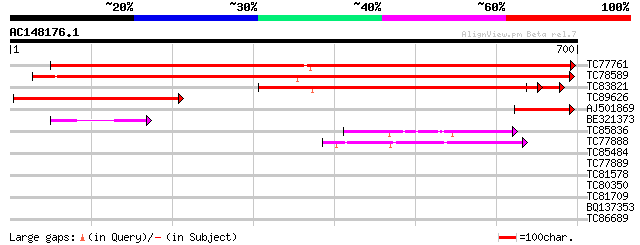
Score E
Sequences producing significant alignments: (bits) Value
TC77761 1-deoxy-D-xylulose 5-phosphate synthase 1 [Medicago trun... 791 0.0
TC78589 1-deoxy-D-xylulose 5-phosphate synthase 2 [Medicago trun... 729 0.0
TC83821 similar to GP|8953400|emb|CAB96673.1 1-D-deoxyxylulose 5... 684 0.0
TC89626 similar to GP|5803266|dbj|BAA83576.1 ESTs AU078063(S1549... 422 e-118
AJ501869 homologue to GP|21322715|emb 1-deoxy-D-xylulose 5-phosp... 97 2e-20
BE321373 GP|21322713|e 1-deoxy-D-xylulose 5-phosphate synthase 1... 85 1e-16
TC85836 homologue to SP|P52904|ODPB_PEA Pyruvate dehydrogenase E... 62 1e-09
TC77888 homologue to PIR|E84758|E84758 probable pyruvate dehydro... 52 1e-06
TC85484 similar to PIR|S58083|S58083 transketolase (EC 2.2.1.1) ... 31 1.4
TC77889 homologue to GP|13605515|gb|AAK32751.1 At1g30120/T2H7_8 ... 30 3.1
TC81578 GP|7688063|emb|CAB89693.1 constitutively photomorphogeni... 30 4.1
TC80350 similar to GP|21539537|gb|AAM53321.1 unknown protein {Ar... 30 4.1
TC81709 similar to PIR|T05867|T05867 hypothetical protein T29A15... 29 5.4
BQ137353 similar to GP|11544433|em dJ1182A14.1 (similar to rat E... 29 7.0
TC86689 similar to GP|17381244|gb|AAL36041.1 AT3g23600/MDB19_9 {... 29 7.0
>TC77761 1-deoxy-D-xylulose 5-phosphate synthase 1 [Medicago truncatula]
Length = 2595
Score = 791 bits (2044), Expect = 0.0
Identities = 409/662 (61%), Positives = 499/662 (74%), Gaps = 14/662 (2%)
Frame = +3
Query: 51 SRPDIDDFYWEKVPTPILDTVQNPLCLKNLSQQELKQLAAEIRLELSSILSGTQILLNPS 110
S ++ ++Y ++ PTP+LDT+ P+ +KNLS +ELKQLA E+R ++ +S T L S
Sbjct: 294 SLSEMGEYYSQRPPTPLLDTINYPIHMKNLSTKELKQLADELRSDVIFSVSRTGGHLGSS 473
Query: 111 MAVVDLTVAIHHVFHAPVDKILWDVGDQTYAHKILTGRRSLMKTIRKKNGLSGFTSRFES 170
+ VV+LT+A+H+VF+ P DKILWDVG Q+Y HKILTGRR M T+R+ NGLSGFT R ES
Sbjct: 474 LGVVELTIALHYVFNTPQDKILWDVGHQSYPHKILTGRRDKMHTMRQTNGLSGFTKRSES 653
Query: 171 EYDAFGAGHGCNSISAGLGMAVARDIKGRRERVVAVISNWTTMSGQVYEAMSNAGYLDSN 230
EYD+FG GH +ISAGLGMAV RD+KGR+ VVAVI + +GQ YEAM+NAGYLDS+
Sbjct: 654 EYDSFGTGHSSTTISAGLGMAVGRDLKGRKNDVVAVIGDGAMTAGQAYEAMNNAGYLDSD 833
Query: 231 LVVILNDSRHSLLPKIE-DGSKTSVNALSSTLSRLQSSKSFRKFREAAKGVTKRIGRGMH 289
++VILND++ LP DG V ALSS LSRLQS+K R+ RE AKGVTKRIG MH
Sbjct: 834 MIVILNDNKQVSLPTATLDGPIPPVGALSSALSRLQSNKPLRELREVAKGVTKRIGGPMH 1013
Query: 290 ELAAKVDEYARGMMGPPGSTLFEELGLYYIGPVDGHNIEDLISVLQEVASLDSMGPVLIH 349
ELAAKVDEYARGM+ GSTLFEELGLYYIGPVDGHNI+DL+++L+EV + +S GPVLIH
Sbjct: 1014ELAAKVDEYARGMISGTGSTLFEELGLYYIGPVDGHNIDDLVAILKEVKTTNSTGPVLIH 1193
Query: 350 VITNENQVEEHNKKSYMTDK------------QQDDNAGRLQTYGDCFVESLVAEAEKDK 397
VIT + + + +K+ DK +Q A + Q+Y F E+L+AEA+ DK
Sbjct: 1194VITEKGRGYPYAQKA--ADKYHGVAKFDPPTGKQFKVAAKTQSYTTYFAEALIAEAKADK 1367
Query: 398 DIVVVHAGITTEPSLKLFMEKFPDRIFNVGIAEQHAVTFASGLSCGGLKPFCIIPSSFLQ 457
DI+ +HA + + +F +FP R F+VGIAEQHAVTFA+GL+C GLKPFC I SSFLQ
Sbjct: 1368DIIAIHAAMGGGTGMNIFHRRFPTRCFDVGIAEQHAVTFAAGLACEGLKPFCAIYSSFLQ 1547
Query: 458 RAYDQVVHDVDQQKVPVRFVITSAGLVGSDGPLQCGAFDITFMSCLPNMIVMAPSDEAEL 517
RAYDQVVHDVD QK+PVRF + AGLVGSDGP G+FD+TFM+CLPNM+VMAPSDEAEL
Sbjct: 1548RAYDQVVHDVDLQKLPVRFAMDRAGLVGSDGPTHSGSFDVTFMACLPNMVVMAPSDEAEL 1727
Query: 518 VHMVATAAHINDQPVCFRYPRGALVGKD-EAILDGIPIEIGKGRILVEGKDVALLGYGSM 576
HMVATAA I+D+P CFRYPRG +G + GIP+EIGKGRIL+EG+ VALLGYGS
Sbjct: 1728CHMVATAAAIDDRPSCFRYPRGNGIGVELPTEYKGIPLEIGKGRILIEGERVALLGYGSA 1907
Query: 577 VQNCLKAYSLLANLGIEVTVADARFCKPLDIELLRQLCKHHSFLITVEEGSIGGFGSHVA 636
VQNCL A SL+ G+ +TVADARFCKPLD L+R L K H LITVEEGSIGGFGSHVA
Sbjct: 1908VQNCLAAASLVEQHGLRLTVADARFCKPLDRSLIRSLAKSHEVLITVEEGSIGGFGSHVA 2087
Query: 637 QFIALDGLLDRRIKWRPIVLPDSYIEHASPNQQLNQAGLTGHHIAATALSLLGRTREALS 696
QF+ALDGLLD +KWRP+VLPD YI+H SP QL AGLT HIAAT ++LG+TREAL
Sbjct: 2088QFMALDGLLDGNLKWRPVVLPDRYIDHGSPADQLCMAGLTPSHIAATVFNILGQTREALE 2267
Query: 697 FM 698
M
Sbjct: 2268VM 2273
>TC78589 1-deoxy-D-xylulose 5-phosphate synthase 2 [Medicago truncatula]
Length = 2760
Score = 729 bits (1883), Expect = 0.0
Identities = 380/682 (55%), Positives = 492/682 (71%), Gaps = 13/682 (1%)
Frame = +3
Query: 29 SQFPLSRITYSSSSRILIHRVCSRPDIDDFYWEKVPTPILDTVQNPLCLKNLSQQELKQL 88
+QF + + S R +I + I+ F EK PTP+LDTV P+ +KNL+ ++L+QL
Sbjct: 279 NQFCVMASSSSDGERTIIRKEKDEWKIN-FSAEKPPTPLLDTVNFPVHMKNLTTEDLEQL 455
Query: 89 AAEIRLELSSILSGTQILLNPSMAVVDLTVAIHHVFHAPVDKILWDVGDQTYAHKILTGR 148
AAE+R ++ +S T L+ S+ VV+L+VA+HHVF P DKI+WDVG Q Y HKILTGR
Sbjct: 456 AAELRADIVHSVSDTGGHLSSSLGVVELSVALHHVFDTPDDKIIWDVGHQAYPHKILTGR 635
Query: 149 RSLMKTIRKKNGLSGFTSRFESEYDAFGAGHGCNSISAGLGMAVARDIKGRRERVVAVIS 208
RS M TIRK +GL+GF R ES +DAFG GH SISAGLGMA+ARD+ G++ V++VI
Sbjct: 636 RSRMHTIRKTSGLAGFPKRDESVHDAFGVGHSSTSISAGLGMAIARDLLGKKNSVISVIG 815
Query: 209 NWTTMSGQVYEAMSNAGYLDSNLVVILNDSRHSLLPKIE-DGSKTSVNALSSTLSRLQSS 267
+ +GQ YEAM+NAG++DSNL+VILND++ LP DG T V ALSSTLS++Q+S
Sbjct: 816 DGAMTAGQAYEAMNNAGFIDSNLIVILNDNKQVSLPTATLDGPATPVGALSSTLSKIQAS 995
Query: 268 KSFRKFREAAKGVTKRIGRGMHELAAKVDEYARGMMGPPGSTLFEELGLYYIGPVDGHNI 327
+ FRK REA K +TK+IG H +A+KVD+YAR + GS+LFEELG+YYIGP+DGHNI
Sbjct: 996 RKFRKLREATKNITKQIGGQTHLVASKVDKYARDFISGSGSSLFEELGMYYIGPMDGHNI 1175
Query: 328 EDLISVLQEVASLDSMGPVLIHVITNE-----------NQVEEHNKKSYMTDKQQDDNAG 376
EDL+++ ++V ++ + GPVLIH++T + +++ K T Q
Sbjct: 1176EDLVNIFEKVKAMPAPGPVLIHIVTEKGKGYPPALAAADRMHGVVKFDPKTGHQFKPKPS 1355
Query: 377 RLQTYGDCFVESLVAEAEKDKDIVVVHAGITTEPSLKLFMEKFPDRIFNVGIAEQHAVTF 436
L Y F +SL+ EAE D IV +HA + L F ++FPDR F+VGIAEQHAVTF
Sbjct: 1356TL-AYTQYFADSLIKEAEMDNKIVAIHAAMGGGTGLNYFQKRFPDRCFDVGIAEQHAVTF 1532
Query: 437 ASGLSCGGLKPFCIIPSSFLQRAYDQVVHDVDQQKVPVRFVITSAGLVGSDGPLQCGAFD 496
A+GL+ GLKPFC I SSFLQR YDQVVHDVD QK+PVRF + AGLVG+DGP CGAFD
Sbjct: 1533AAGLAAEGLKPFCAIYSSFLQRGYDQVVHDVDLQKLPVRFAMDRAGLVGADGPTHCGAFD 1712
Query: 497 ITFMSCLPNMIVMAPSDEAELVHMVATAAHINDQPVCFRYPRGALVGKDEAILD-GIPIE 555
ITFM+CLPNMIVMAPSDEAEL++MVATAA I+D+P CFR+PRG +G + + + G P+E
Sbjct: 1713ITFMACLPNMIVMAPSDEAELMNMVATAAAIDDRPSCFRFPRGNGIGANLPLNNKGTPLE 1892
Query: 556 IGKGRILVEGKDVALLGYGSMVQNCLKAYSLLANLGIEVTVADARFCKPLDIELLRQLCK 615
IGKGRIL+EG VA+LGYG MVQ C+KA +L +G+ VTVADARFCKPLD +L+R L +
Sbjct: 1893IGKGRILLEGSRVAILGYGCMVQQCMKAAEMLRAVGVYVTVADARFCKPLDTDLIRLLAR 2072
Query: 616 HHSFLITVEEGSIGGFGSHVAQFIALDGLLDRRIKWRPIVLPDSYIEHASPNQQLNQAGL 675
H LITVEEGSIGGFGSHV+QF++L GLLD +K R ++LPD YI+H +PN Q+++AGL
Sbjct: 2073EHEILITVEEGSIGGFGSHVSQFLSLAGLLDGPLKLRSMMLPDRYIDHGAPNDQIDEAGL 2252
Query: 676 TGHHIAATALSLLGRTREALSF 697
+ HI AT LSLL +EAL F
Sbjct: 2253SSKHILATVLSLLDMPKEALLF 2318
>TC83821 similar to GP|8953400|emb|CAB96673.1 1-D-deoxyxylulose 5-phosphate
synthase-like protein {Arabidopsis thaliana}, partial
(46%)
Length = 1172
Score = 684 bits (1765), Expect = 0.0
Identities = 347/359 (96%), Positives = 348/359 (96%), Gaps = 9/359 (2%)
Frame = +1
Query: 308 STLFEELGLYYIGPVDGHNIEDLISVLQEVASLDSMGPVLIHVITNENQVEEHNKKSYMT 367
STLFEELGLYYIGPVDGHNIEDLISVLQEVASLDSMGPVLIHVITNENQVEEHNKKSYMT
Sbjct: 1 STLFEELGLYYIGPVDGHNIEDLISVLQEVASLDSMGPVLIHVITNENQVEEHNKKSYMT 180
Query: 368 DKQQD--------DNAGRLQTYGDCFVESLVAEAEKDKDIVVVHAGITTEPSLKLFMEKF 419
DKQQD DNAGRLQTYGDCFVESLVAEAEKDKDIVVVHAGITTEPSLKLFMEKF
Sbjct: 181 DKQQDESVSFDLLDNAGRLQTYGDCFVESLVAEAEKDKDIVVVHAGITTEPSLKLFMEKF 360
Query: 420 PDRIFNVGIAEQHAVTFASGLSCGGLKPFCIIPSSFLQRAYDQVVHDVDQQKVPVRFVIT 479
PDRIFNVGIAEQHAVTFASGLSCGGLKPFCIIPSSFLQRAYDQVVHDVDQQKVPVRFVIT
Sbjct: 361 PDRIFNVGIAEQHAVTFASGLSCGGLKPFCIIPSSFLQRAYDQVVHDVDQQKVPVRFVIT 540
Query: 480 SAGLVGSDGPLQCGAFDITFMSCLPNMIVMAPSDEAELVHMVATAAHINDQPVCFRYPRG 539
SA LVGSDGPLQCGAFDITFMSCLPNMIVMAPSDEAELVHMVATAAHINDQPVCFRYPRG
Sbjct: 541 SARLVGSDGPLQCGAFDITFMSCLPNMIVMAPSDEAELVHMVATAAHINDQPVCFRYPRG 720
Query: 540 ALVGKDEAILDGIPIEIGKGRILVEGKDVALLGYGSMVQNCLKAYSLLANLGIEVTVADA 599
ALVGKDEAILDGIPIEIGKGRILVEGKDVALLGYGSMVQNCLKAYSLLANLGIEVTVADA
Sbjct: 721 ALVGKDEAILDGIPIEIGKGRILVEGKDVALLGYGSMVQNCLKAYSLLANLGIEVTVADA 900
Query: 600 RFCKPLDIELLRQLCKHHSFLITVEEGSIGGFGSHVAQFIALDGLLDR-RIKWRPIVLP 657
RFCKPLDIELLRQLCKHHSFLITVEEGSIGGFGSHVAQFIALDGLLDR + KWRPIVLP
Sbjct: 901 RFCKPLDIELLRQLCKHHSFLITVEEGSIGGFGSHVAQFIALDGLLDRKKFKWRPIVLP 1077
Score = 58.5 bits (140), Expect = 8e-09
Identities = 27/47 (57%), Positives = 34/47 (71%)
Frame = +2
Query: 639 IALDGLLDRRIKWRPIVLPDSYIEHASPNQQLNQAGLTGHHIAATAL 685
+ +D +R + + DSYIEHA PNQQLNQAGLTGHHIAAT++
Sbjct: 1022 LLMDFSTERNLSGDQLSYQDSYIEHALPNQQLNQAGLTGHHIAATSI 1162
>TC89626 similar to GP|5803266|dbj|BAA83576.1 ESTs AU078063(S15496)
C97608(C60475) C28255(C60475) correspond to a region of
the predicted gene.~, partial (24%)
Length = 748
Score = 422 bits (1085), Expect = e-118
Identities = 210/210 (100%), Positives = 210/210 (100%)
Frame = +3
Query: 5 VCARYPFVIPFHSQTLTRRLDCSISQFPLSRITYSSSSRILIHRVCSRPDIDDFYWEKVP 64
VCARYPFVIPFHSQTLTRRLDCSISQFPLSRITYSSSSRILIHRVCSRPDIDDFYWEKVP
Sbjct: 117 VCARYPFVIPFHSQTLTRRLDCSISQFPLSRITYSSSSRILIHRVCSRPDIDDFYWEKVP 296
Query: 65 TPILDTVQNPLCLKNLSQQELKQLAAEIRLELSSILSGTQILLNPSMAVVDLTVAIHHVF 124
TPILDTVQNPLCLKNLSQQELKQLAAEIRLELSSILSGTQILLNPSMAVVDLTVAIHHVF
Sbjct: 297 TPILDTVQNPLCLKNLSQQELKQLAAEIRLELSSILSGTQILLNPSMAVVDLTVAIHHVF 476
Query: 125 HAPVDKILWDVGDQTYAHKILTGRRSLMKTIRKKNGLSGFTSRFESEYDAFGAGHGCNSI 184
HAPVDKILWDVGDQTYAHKILTGRRSLMKTIRKKNGLSGFTSRFESEYDAFGAGHGCNSI
Sbjct: 477 HAPVDKILWDVGDQTYAHKILTGRRSLMKTIRKKNGLSGFTSRFESEYDAFGAGHGCNSI 656
Query: 185 SAGLGMAVARDIKGRRERVVAVISNWTTMS 214
SAGLGMAVARDIKGRRERVVAVISNWTTMS
Sbjct: 657 SAGLGMAVARDIKGRRERVVAVISNWTTMS 746
>AJ501869 homologue to GP|21322715|emb 1-deoxy-D-xylulose 5-phosphate
synthase 2 {Medicago truncatula}, partial (10%)
Length = 394
Score = 97.4 bits (241), Expect = 2e-20
Identities = 46/74 (62%), Positives = 59/74 (79%)
Frame = +2
Query: 624 EEGSIGGFGSHVAQFIALDGLLDRRIKWRPIVLPDSYIEHASPNQQLNQAGLTGHHIAAT 683
EEGSIGGFGSHV+QF++L GLLD +K R ++LPD YI+H +PN Q+++AGL+ HI AT
Sbjct: 44 EEGSIGGFGSHVSQFLSLAGLLDGPLKLRSMMLPDRYIDHGAPNDQIDEAGLSSKHILAT 223
Query: 684 ALSLLGRTREALSF 697
LSLL T+EAL F
Sbjct: 224 VLSLLDMTKEALLF 265
>BE321373 GP|21322713|e 1-deoxy-D-xylulose 5-phosphate synthase 1 {Medicago
truncatula}, partial (18%)
Length = 399
Score = 84.7 bits (208), Expect = 1e-16
Identities = 48/125 (38%), Positives = 63/125 (50%)
Frame = +1
Query: 51 SRPDIDDFYWEKVPTPILDTVQNPLCLKNLSQQELKQLAAEIRLELSSILSGTQILLNPS 110
S ++ ++Y ++ PTP+LDT+ P+ +KNLS +
Sbjct: 160 SLSEMGEYYSQRPPTPLLDTINYPIHMKNLSTK--------------------------- 258
Query: 111 MAVVDLTVAIHHVFHAPVDKILWDVGDQTYAHKILTGRRSLMKTIRKKNGLSGFTSRFES 170
DKILWDVG Q+Y HKILTGRR M T+R+ NGLSGFT R ES
Sbjct: 259 ------------------DKILWDVGHQSYPHKILTGRRDKMHTMRQTNGLSGFTKRSES 384
Query: 171 EYDAF 175
EYD+F
Sbjct: 385 EYDSF 399
>TC85836 homologue to SP|P52904|ODPB_PEA Pyruvate dehydrogenase E1 component
beta subunit mitochondrial precursor (EC 1.2.4.1)
(PDHE1-B)., complete
Length = 1507
Score = 61.6 bits (148), Expect = 1e-09
Identities = 63/229 (27%), Positives = 96/229 (41%), Gaps = 15/229 (6%)
Frame = +2
Query: 413 KLFMEKF-PDRIFNVGIAEQHAVTFASGLSCGGLKPFC-IIPSSFLQRAYDQVVHDV--- 467
K +EK+ P+R+ + I E G + GLKP + +F +A D +++
Sbjct: 308 KGLLEKYGPERVLDTPITEAGFTGIGVGAAYYGLKPVVEFMTFNFSMQAIDHIINSAAKS 487
Query: 468 -----DQQKVPVRFVITSAGLVGSDGPLQCGAFDITFMSCLPNMIVMAPSDEAELVHMVA 522
Q VP+ F + G G + + SC P + V+AP + ++
Sbjct: 488 NYMSAGQINVPIVFRGPNGAAAGV-GAQHSHCYASWYGSC-PGLKVLAPYSSEDARGLLK 661
Query: 523 TAAHINDQPVCFRYPRGALVGKD-----EAILDGIPIEIGKGRILVEGKDVALLGYGSMV 577
A D PV F L G+ E + + IGK +I EGKDV + + MV
Sbjct: 662 AAIRDPD-PVVF-LENELLYGESFPVSAEVLDSSFCLPIGKAKIEREGKDVTITAFSKMV 835
Query: 578 QNCLKAYSLLANLGIEVTVADARFCKPLDIELLRQLCKHHSFLITVEEG 626
LKA L GI V + R +PLD + + + L+TVEEG
Sbjct: 836 GFALKAAETLEKEGISAEVINLRSIRPLDRATINASVRKTNRLVTVEEG 982
>TC77888 homologue to PIR|E84758|E84758 probable pyruvate dehydrogenase E1
beta subunit [imported] - Arabidopsis thaliana, partial
(85%)
Length = 1707
Score = 51.6 bits (122), Expect = 1e-06
Identities = 67/270 (24%), Positives = 113/270 (41%), Gaps = 17/270 (6%)
Frame = +2
Query: 387 ESLVAEAEKDKDIVVV-----HAGITTEPSLKLFMEKFPD-RIFNVGIAEQHAVTFASGL 440
E L E E+D + V+ H G + + + L EKF D R+ + IAE G
Sbjct: 455 EGLEEEMERDPCVCVMGEDVGHYGGSYKVTRNL-AEKFGDLRVLDTPIAENAFTGMGIGA 631
Query: 441 SCGGLKPFCI-IPSSFLQRAYDQVVHDVD--------QQKVPVRFVITSAGLVGSDGPLQ 491
+ GL+P + FL A++Q+ ++ Q K+PV VI G VG +
Sbjct: 632 AMTGLRPIIEGMNMGFLLLAFNQISNNCGMLHYTSGGQFKIPV--VIRGPGGVGRQLGAE 805
Query: 492 CGAFDITFMSCLPNMIVMAPSDEAELVHMVATAAHINDQPVCFRYPRGALVGKDEAILDG 551
++ +P + ++A S ++ A ++ + F + L E I D
Sbjct: 806 HSQRLESYFQSIPGIQMVACSTPYNAKGLMKAAIRSDNPVILFEHV--LLYNLKERIPDE 979
Query: 552 -IPIEIGKGRILVEGKDVALLGYGSMVQNCLKAYSLLANLGIEVTVADARFCKPLDIELL 610
+ + + ++ G+ V +L Y M + ++A L N G + V D R KP D+ +
Sbjct: 980 EYVLSLEEAEMVRPGEHVTILTYSRMRYHVMQAAKTLVNKGYDPEVIDIRSLKPFDLHTI 1159
Query: 611 -RQLCKHHSFLITVEEGSIGGFGSHVAQFI 639
+ K H LI E GG G+ + I
Sbjct: 1160GNSIKKTHRVLIVEECMRTGGIGASLTAAI 1249
>TC85484 similar to PIR|S58083|S58083 transketolase (EC 2.2.1.1) precursor -
potato (fragment), partial (95%)
Length = 2685
Score = 31.2 bits (69), Expect = 1.4
Identities = 32/130 (24%), Positives = 51/130 (38%), Gaps = 7/130 (5%)
Frame = +1
Query: 474 VRFVITSAGL-VGSDGPLQCGAFDITFMSCLPNMIVMAPSDEAELVHMVATAAHINDQPV 532
V +V+T + +G DGP + +PN++++ P+D E A +P
Sbjct: 1675 VIYVMTHDSIGLGEDGPTHQPIEHLASFRAMPNILMLRPADGNETAGAYKVAVLNRKRPS 1854
Query: 533 CFRYPRGALVGKDEAILDGIPIEIGKGRILVEGK------DVALLGYGSMVQNCLKAYSL 586
R L ++G+ KG +V DV L+G GS ++ KA
Sbjct: 1855 ILALSRQKLPNLAGTSIEGVE----KGGYIVSDNSTGNKPDVILIGTGSELEIAYKAGED 2022
Query: 587 LANLGIEVTV 596
L G V V
Sbjct: 2023 LRKEGKAVRV 2052
>TC77889 homologue to GP|13605515|gb|AAK32751.1 At1g30120/T2H7_8
{Arabidopsis thaliana}, partial (28%)
Length = 653
Score = 30.0 bits (66), Expect = 3.1
Identities = 20/65 (30%), Positives = 29/65 (43%), Gaps = 1/65 (1%)
Frame = +3
Query: 576 MVQNCLKAYSLLANLGIEVTVADARFCKPLDIELL-RQLCKHHSFLITVEEGSIGGFGSH 634
M + ++A L N G + V D R KP D+ + + K H LI E GG G+
Sbjct: 6 MRYHVMQAAKTLVNKGYDPEVIDIRSLKPFDLHTIGNSIKKTHRVLIVEECMRTGGIGAS 185
Query: 635 VAQFI 639
+ I
Sbjct: 186 LTAAI 200
>TC81578 GP|7688063|emb|CAB89693.1 constitutively photomorphogenic 1 protein
{Pisum sativum}, partial (39%)
Length = 842
Score = 29.6 bits (65), Expect = 4.1
Identities = 18/61 (29%), Positives = 31/61 (50%), Gaps = 3/61 (4%)
Frame = +2
Query: 336 EVASLDSMGPVLIHVITNENQV---EEHNKKSYMTDKQQDDNAGRLQTYGDCFVESLVAE 392
++AS D G V + +T + EEH K+++ D + D + + DC V+ LV E
Sbjct: 401 QIASSDYEGIVTVWDVTTRKSLMEYEEHEKRAWSVDFSRTDPSMLVSGSDDCKVKVLVHE 580
Query: 393 A 393
+
Sbjct: 581 S 583
>TC80350 similar to GP|21539537|gb|AAM53321.1 unknown protein {Arabidopsis
thaliana}, partial (42%)
Length = 1175
Score = 29.6 bits (65), Expect = 4.1
Identities = 19/59 (32%), Positives = 30/59 (50%), Gaps = 1/59 (1%)
Frame = -2
Query: 573 YGSMVQNCLKA-YSLLANLGIEVTVADARFCKPLDIELLRQLCKHHSFLITVEEGSIGG 630
+GS++ C + YSLL+ L + A +DI L +QLCK + T+E + G
Sbjct: 193 FGSLIFCCNAS*YSLLSKLLAPLNTATTELILEVDIALRKQLCKT*NTSSTIEVVRLSG 17
>TC81709 similar to PIR|T05867|T05867 hypothetical protein T29A15.110 -
Arabidopsis thaliana, partial (72%)
Length = 1383
Score = 29.3 bits (64), Expect = 5.4
Identities = 20/74 (27%), Positives = 29/74 (39%)
Frame = -3
Query: 60 WEKVPTPILDTVQNPLCLKNLSQQELKQLAAEIRLELSSILSGTQILLNPSMAVVDLTVA 119
W K P + + +P CL ++S IR S + S LL+PS L
Sbjct: 1147 WSKYSFPSIASFNSPTCLFSIS----------IRSSSSFVFSWVSKLLSPSRCPFGLPFL 998
Query: 120 IHHVFHAPVDKILW 133
+ PVD + W
Sbjct: 997 VSTDQAVPVDDLSW 956
>BQ137353 similar to GP|11544433|em dJ1182A14.1 (similar to rat Espin) {Homo
sapiens}, partial (2%)
Length = 1195
Score = 28.9 bits (63), Expect = 7.0
Identities = 16/32 (50%), Positives = 20/32 (62%), Gaps = 1/32 (3%)
Frame = +1
Query: 21 TRRLDCSISQFP-LSRITYSSSSRILIHRVCS 51
TR++ CS SQ L + Y SSRIL + VCS
Sbjct: 997 TRQITCSCSQHTRLYSVLYD*SSRILSYHVCS 1092
>TC86689 similar to GP|17381244|gb|AAL36041.1 AT3g23600/MDB19_9 {Arabidopsis
thaliana}, partial (96%)
Length = 1264
Score = 28.9 bits (63), Expect = 7.0
Identities = 15/52 (28%), Positives = 25/52 (47%)
Frame = -2
Query: 570 LLGYGSMVQNCLKAYSLLANLGIEVTVADARFCKPLDIELLRQLCKHHSFLI 621
L G SM+ L+ ++ + + + + F KP+ L+ LC HSF I
Sbjct: 978 LCGMSSMLHGSLEVHTWIQRISTDKYFSLEMFSKPVQ*VLMCLLCSFHSFCI 823
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.137 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,579,354
Number of Sequences: 36976
Number of extensions: 278608
Number of successful extensions: 1210
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 1195
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1202
length of query: 700
length of database: 9,014,727
effective HSP length: 103
effective length of query: 597
effective length of database: 5,206,199
effective search space: 3108100803
effective search space used: 3108100803
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148176.1


