

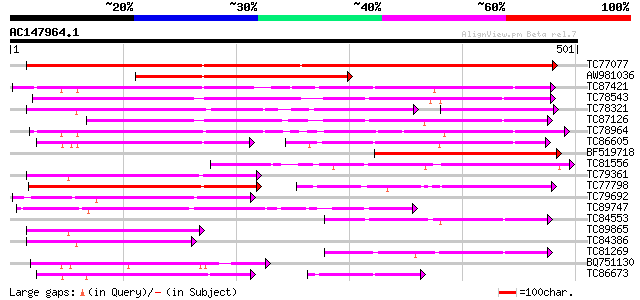
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147964.1 + phase: 0
(501 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77077 similar to GP|9293909|dbj|BAB01812.1 sugar transporter p... 582 e-166
AW981036 weakly similar to PIR|G84564|G84 probable sugar transpo... 231 5e-61
TC87421 sugar transporter 203 1e-52
TC78543 similar to PIR|T14545|T14545 probable sugar transporter ... 203 1e-52
TC78321 similar to GP|21689841|gb|AAM67564.1 unknown protein {Ar... 189 2e-48
TC87126 similar to GP|15724240|gb|AAL06513.1 At1g75220/F22H5_6 {... 187 6e-48
TC78964 weakly similar to PIR|T00450|T00450 probable monosacchar... 187 1e-47
TC86605 similar to PIR|T01853|T01853 probable hexose transport p... 110 3e-46
BF519718 weakly similar to PIR|G84564|G84 probable sugar transpo... 180 1e-45
TC81556 similar to GP|2688830|gb|AAB88879.1| putative sugar tran... 147 9e-36
TC79361 similar to GP|17380890|gb|AAL36257.1 putative membrane t... 140 9e-34
TC77798 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_14... 138 6e-33
TC79692 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_14... 126 2e-29
TC89747 similar to PIR|F96696|F96696 protein F1N21.12 [imported]... 115 3e-26
TC84553 similar to GP|17381265|gb|AAL36051.1 AT5g18840/F17K4_90 ... 112 3e-25
TC89865 similar to GP|17380890|gb|AAL36257.1 putative membrane t... 105 5e-23
TC84386 similar to GP|21689841|gb|AAM67564.1 unknown protein {Ar... 100 1e-21
TC81269 similar to GP|23504385|emb|CAC00697. putative sugar tran... 100 2e-21
BQ751130 weakly similar to GP|15289796|dbj putative monosacchari... 100 2e-21
TC86673 weakly similar to PIR|S25015|S25015 monosaccharide trans... 99 5e-21
>TC77077 similar to GP|9293909|dbj|BAB01812.1 sugar transporter protein
{Arabidopsis thaliana}, partial (85%)
Length = 1752
Score = 582 bits (1500), Expect = e-166
Identities = 289/469 (61%), Positives = 367/469 (77%)
Frame = +1
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLT 75
NKYAFACA++ASM SI+ GYD GVMSGA ++IK DL +SD + EVL GI+N+ +L+GS
Sbjct: 100 NKYAFACAMLASMTSILLGYDIGVMSGAAIYIKRDLKVSDGKIEVLLGIINIYSLIGSCL 279
Query: 76 AGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSA 135
AGRTSD+IGRRYTI A +F +GA LMG+ PNY LM GR V GVG+G+ALMIAPVY+A
Sbjct: 280 AGRTSDWIGRRYTIVFAGAIFFVGALLMGFSPNYNFLMFGRFVAGVGIGYALMIAPVYTA 459
Query: 136 EISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFC 195
E+S ASSRGFLTS PE+ I GILLGYISNY K LSLKLGWR+MLG+ A+PS ++A
Sbjct: 460 EVSPASSRGFLTSFPEVFINSGILLGYISNYAFSK-LSLKLGWRMMLGVGAIPSVILAVG 636
Query: 196 ILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQ 255
+L MPESPRWLVM+G+LG A KVL + S++ +EA+LRL +IK AAG+ E+CND+ V++
Sbjct: 637 VLAMPESPRWLVMRGRLGDAIKVLNKTSDSKEEAQLRLAEIKQAAGIPEDCNDDVVEVKV 816
Query: 256 KSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEK 315
K+ GEGVWKEL L PTP+VR ++IAA+GIHFF+ A+G++AV+LYSP IF+KAGI
Sbjct: 817 KN-TGEGVWKELFLYPTPAVRHIVIAALGIHFFQQASGVDAVVLYSPTIFKKAGINGDTH 993
Query: 316 LLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNV 375
LL+ATI VG K +F+++A F+LD+ GRR LL S GGM++ L L +SLT++D SN +
Sbjct: 994 LLIATIAVGFVKTLFILVATFMLDRYGRRPLLLTSVGGMVVSLLTLAVSLTIIDHSNTKL 1173
Query: 376 LWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMT 435
WA+ LSI +YVA F+IG GPITWVYSSEIFPL+LRAQGA+ GV VNR + V+SMT
Sbjct: 1174NWAIGLSIATVLSYVATFSIGAGPITWVYSSEIFPLRLRAQGAACGVVVNRVTSGVISMT 1353
Query: 436 FISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFTK 484
F+S+ K ITIGG+FF+F GI+ + W+FFY LPET+GK LEEME F K
Sbjct: 1354FLSLSKGITIGGAFFLFGGIATIGWIFFYIMLPETQGKTLEEMEASFGK 1500
>AW981036 weakly similar to PIR|G84564|G84 probable sugar transporter
[imported] - Arabidopsis thaliana, partial (25%)
Length = 575
Score = 231 bits (589), Expect = 5e-61
Identities = 113/192 (58%), Positives = 145/192 (74%)
Frame = +2
Query: 112 LMVGRCVCGVGVGFALMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKY 171
LM+GRC+ G GVGFAL+I VYSAEISS S RGFLTSLP+L I IG LLGY+SNY LGK
Sbjct: 2 LMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGK- 178
Query: 172 LSLKLGWRLMLGIAALPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAEL 231
LSL+LGWR+ML I ++PS + +L + ESPRWLVMQG+LG AKKVL+ +SN+ QEAE
Sbjct: 179 LSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQ 358
Query: 232 RLKDIKIAAGLDENCNDETVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHA 291
R+K+IK A G+DENC V + +K+ G G KE+ +P+P V +LIAA+G+H F++
Sbjct: 359 RMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNI 538
Query: 292 TGIEAVMLYSPR 303
G+E + LYSPR
Sbjct: 539 CGVEGIFLYSPR 574
>TC87421 sugar transporter
Length = 1753
Score = 203 bits (517), Expect = 1e-52
Identities = 151/504 (29%), Positives = 241/504 (46%), Gaps = 24/504 (4%)
Frame = +2
Query: 3 INQGDKEDQTNTFNKYAFACAIVASMVSIVSGYDTGVMSGAM------------LFIKED 50
I G+KE N C IVA+M ++ GYD G+ G ++ K++
Sbjct: 50 IGGGNKEYPGNLTPFVTITC-IVAAMGGLIFGYDIGISGGVTSMDPFLKKFFPAVYRKKN 226
Query: 51 LGISDTQQ--------EVLAGILNLCALVGSLTAGRTSDYIGRRYTIFLASILFILGAGL 102
S Q + L L AL+ SL A + GR+ ++ +LF++GA +
Sbjct: 227 KDKSTNQYCQYDSQTLTMFTSSLYLAALLSSLVASTITRRFGRKLSMLFGGLLFLVGALI 406
Query: 103 MGYGPNYAILMVGRCVCGVGVGFALMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGY 162
G+ + +L+VGR + G G+GFA P+Y +E++ RG L +L I IGIL+
Sbjct: 407 NGFANHVWMLIVGRILLGFGIGFANQPVPLYLSEMAPYKYRGALNIGFQLSITIGILVAN 586
Query: 163 ISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQV 222
+ NY K + GWRL LG A +P+ ++ L +P++P ++ +G AK
Sbjct: 587 VLNYFFAK-IKGGWGWRLSLGGAMVPALIITIGSLVLPDTPNSMIERGDRDGAK------ 745
Query: 223 SNTTQEAELRLKDIKIAAGLDENCNDETVKLPQKSHQGEGVWKELILRPTPSVRWMLIAA 282
+LK I+ +DE ND V + S Q E W+ L+ R R L A
Sbjct: 746 --------AQLKRIRGIEDVDEEFND-LVAASEASMQVENPWRNLLQR---KYRPQLTMA 889
Query: 283 VGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLG 342
V I FF+ TGI +M Y+P +F G L+ A I G+ +V ++++ +DK G
Sbjct: 890 VLIPFFQQFTGINVIMFYAPVLFNSIGFKDDASLMSAVI-TGVVNVVATCVSIYGVDKWG 1066
Query: 343 RRRLLQISTGGMIIGLTLLGLSLTVVDKSNGN----VLWALILSIVATYAYVAFFNIGLG 398
RR L M+I + ++ ++GN W I+ ++ YVA F G
Sbjct: 1067RRALFLEGGAQMLICQVAVAAAIGAKFGTSGNPGNLPEWYAIVVVLFICIYVAGFAWSWG 1246
Query: 399 PITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTFISIYKAITIGGSFFMFAGISVL 458
P+ W+ SEIFPL++R+ S+ V+VN +V+ F+ + + G F FA ++
Sbjct: 1247PLGWLVPSEIFPLEIRSAAQSVNVSVNMLFTFLVAQVFLIMLCHMKF-GLFLFFAFFVLV 1423
Query: 459 AWLFFYFFLPETKGKALEEMEMVF 482
++ +F LPETKG +EEM+ V+
Sbjct: 1424MSIYVFFLLPETKGIPIEEMDRVW 1495
>TC78543 similar to PIR|T14545|T14545 probable sugar transporter protein -
beet, partial (93%)
Length = 1653
Score = 203 bits (517), Expect = 1e-52
Identities = 130/468 (27%), Positives = 239/468 (50%), Gaps = 6/468 (1%)
Frame = +1
Query: 21 ACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLTAGRTS 80
AC ++ ++ I G+ G S I DLG+S ++ + + N+ A+VG++ +G+ +
Sbjct: 154 ACVLIVALGPIQFGFTAGYTSPTQSAIITDLGLSVSEFSLFGSLSNVGAMVGAIASGQIA 333
Query: 81 DYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSAEISSA 140
+Y+GR+ ++ +ASI I+G ++ + + + L +GR + G GVG PVY AEIS
Sbjct: 334 EYMGRKGSLMIASIPNIIGWLMISFANDSSFLYMGRLLEGFGVGIISYTVPVYIAEISPQ 513
Query: 141 SSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFCILTMP 200
+ RG L S+ +L + +GI+L Y+ L L + WR + + +P ++ + +P
Sbjct: 514 NLRGSLVSVNQLSVTLGIMLAYL--------LGLFVEWRFLAILGIIPCTLLIPGLFFIP 669
Query: 201 ESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIA-AGLDENCNDETVKLPQKSHQ 259
ESPRWL G + + L + + + + +IK A A + +L Q+ +
Sbjct: 670 ESPRWLAKMGMTEEFENSLQVLRGFETDISVEVNEIKTAVASANRRTTVRFSELKQRRY- 846
Query: 260 GEGVWKELILRPTPSVRWM-LIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKLLL 318
W+ L+ +G+ + +GI V+ YS IF+ AGI+S + +
Sbjct: 847 -----------------WLPLMIGIGLLVLQQLSGINGVLFYSSTIFQNAGISSSD---V 966
Query: 319 ATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDK--SNGNVL 376
AT GVG +++ + L+L DK GRR LL +S+ M + L ++ +S + D S + L
Sbjct: 967 ATFGVGAVQVLATTLTLWLADKSGRRLLLIVSSSAMTLSLLVVSISFYLKDYYISADSSL 1146
Query: 377 WAL--ILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSM 434
+ + +LS+ V F++G+G + W+ SEI P+ ++ S N + ++++
Sbjct: 1147YGILSLLSVAGVVVMVIAFSLGMGAMPWIIMSEILPINIKGLAGSFATLANWFFSWLITL 1326
Query: 435 TFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVF 482
T ++ + GG+F ++ + F ++PETKGK LEE++ F
Sbjct: 1327T-ANLLLDWSSGGTFTIYTVVCAFTVGFVAIWVPETKGKTLEEIQQFF 1467
>TC78321 similar to GP|21689841|gb|AAM67564.1 unknown protein {Arabidopsis
thaliana}, partial (80%)
Length = 2093
Score = 189 bits (480), Expect = 2e-48
Identities = 119/349 (34%), Positives = 188/349 (53%), Gaps = 3/349 (0%)
Frame = +2
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQ---QEVLAGILNLCALVG 72
N Y A A + ++ GYDTGV+SGA+L+I+++ + + QE + A++G
Sbjct: 212 NPYVLRLAFSAGIGGLLFGYDTGVISGALLYIRDEFPAVEKKTWLQEAIVSTAIAGAIIG 391
Query: 73 SLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPV 132
+ G +D GR+ +I +A LF+LG+ ++ PN A L+VGR G+GVG A M +P+
Sbjct: 392 AAIGGWINDRFGRKVSIIVADTLFLLGSIILAAAPNPATLIVGRVFVGLGVGMASMASPL 571
Query: 133 YSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVV 192
Y +E S RG L SL I G L Y+ N K WR MLG+AA P+ +
Sbjct: 572 YISEASPTRVRGALVSLNSFLITGGQFLSYLINLAFTKAPGT---WRWMLGVAAAPAVIQ 742
Query: 193 AFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVK 252
+L++PESPRWL +G+ +AK +L ++ ++ + ++ +K E+V+
Sbjct: 743 IVLMLSLPESPRWLYRKGKEEEAKVILKKIYE-VEDYDNEIQALK-----------ESVE 886
Query: 253 LPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITS 312
+ K + + + L T SVR L A VG+ FF+ TGI VM YSP I + AG S
Sbjct: 887 MELKETEKISIMQ---LVKTTSVRRGLYAGVGLAFFQQFTGINTVMYYSPSIVQLAGFAS 1057
Query: 313 KEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLL 361
K LL ++ +++++ +DK GR++L IS G+++ LTLL
Sbjct: 1058KRTALLLSLITSGLNAFGSILSIYFIDKTGRKKLALISLTGVVLSLTLL 1204
Score = 78.6 bits (192), Expect = 5e-15
Identities = 40/105 (38%), Positives = 63/105 (59%)
Frame = +2
Query: 381 LSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTFISIY 440
++I+A Y+ FF+ G+G + WV +SEI+PL+ R I N VVS +F+S+
Sbjct: 1490 IAILALALYIIFFSPGMGTVPWVVNSEIYPLRYRGICGGIASTTVWVSNLVVSQSFLSLT 1669
Query: 441 KAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFTKK 485
AI +F +FA I+++A F F+PETKG +EE+E + K+
Sbjct: 1670 VAIGPAWTFMIFAIIAIVAIFFVIIFVPETKGVPMEEVESMLEKR 1804
>TC87126 similar to GP|15724240|gb|AAL06513.1 At1g75220/F22H5_6 {Arabidopsis
thaliana}, partial (79%)
Length = 1524
Score = 187 bits (476), Expect = 6e-48
Identities = 121/414 (29%), Positives = 213/414 (51%), Gaps = 3/414 (0%)
Frame = +3
Query: 69 ALVGSLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALM 128
A+VG++ +G+ ++Y+GR+ ++ +ASI I+G + + + + L +GR + G GVG
Sbjct: 9 AMVGAIASGQIAEYVGRKGSLMIASIPNIIGWLAISFAKDSSFLFMGRLLEGFGVGIISY 188
Query: 129 IAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALP 188
+ PVY AEI+ + RG L S+ +L + IGI+L Y+ L L WR++ + LP
Sbjct: 189 VVPVYIAEIAPENMRGSLGSVNQLSVTIGIMLAYL--------LGLFANWRVLAILGILP 344
Query: 189 SFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCND 248
V+ + +PESPRWL G + + + L + + + + +IK A N
Sbjct: 345 CTVLIPGLFFIPESPRWLAKMGMMEEFETSLQVLRGFDTDISVEVHEIKKAVA----SNG 512
Query: 249 ETVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKA 308
+ + Q + W + L +G+ + +GI V+ YS IF A
Sbjct: 513 KRATIRFADLQRKRYW------------FPLSVGIGLLVLQQLSGINGVLFYSTSIFANA 656
Query: 309 GITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSL--- 365
GI+S AT+G+G +++ +A +L+DK GRR LL IS+ M L ++ ++
Sbjct: 657 GISSSN---AATVGLGAIQVIATGVATWLVDKSGRRVLLIISSSLMTASLLVVSIAFYLE 827
Query: 366 TVVDKSNGNVLWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVN 425
VV+K + I+S+V V F++GLGPI W+ SEI P+ ++ S N
Sbjct: 828 GVVEKDSQYFSILGIISVVGLVVMVIGFSLGLGPIPWLIMSEILPVNIKGLAGSTATMAN 1007
Query: 426 RTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEME 479
+ +++MT ++ + GG+F ++ ++ +F ++PETKG+ LEE++
Sbjct: 1008WLVAWIITMT-ANLLLTWSSGGTFLIYTVVAAFTVVFTSLWVPETKGRTLEEIQ 1166
>TC78964 weakly similar to PIR|T00450|T00450 probable monosaccharide
transport protein T14N5.7 - Arabidopsis thaliana,
partial (98%)
Length = 1771
Score = 187 bits (474), Expect = 1e-47
Identities = 145/499 (29%), Positives = 243/499 (48%), Gaps = 22/499 (4%)
Frame = +1
Query: 18 YAFACAIVASMVSIVSGYDTGVMSGAM------------LFIKEDLGISDTQQ------- 58
+AF C +V ++ + GYD GV G ++ K+ + +T
Sbjct: 106 FAFTC-VVGALGGSLFGYDLGVSGGVTSMDDFLEKFFPDVYRKKHAHLKETDYCKYDNQV 282
Query: 59 -EVLAGILNLCALVGSLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRC 117
+ L ALV + A + GR+ TI + ++ F++GA L N L++GR
Sbjct: 283 LTLFTSSLYFSALVMTFFASYLTRNKGRKATIIVGALSFLIGAILNAAAQNIPTLIIGRV 462
Query: 118 VCGVGVGFALMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLG 177
G G+GF P+Y +E++ ASSRG + L + GIL+ + NY K G
Sbjct: 463 FLGGGIGFGNQAVPLYLSEMAPASSRGAVNQLFQFTTCAGILIANLVNYFTDKI--HPHG 636
Query: 178 WRLMLGIAALPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIK 237
WR+ LG+A +P+ ++ + E+P LV QG+L +A+KVL +V T+ + +D+K
Sbjct: 637 WRISLGLAGIPAVLMLLGGIFCAETPNSLVEQGRLDEARKVLEKVRG-TKNVDAEFEDLK 813
Query: 238 IAAGLDENCNDETVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAV 297
A+ L + VK P K V + RP ++I A+G F+ TG ++
Sbjct: 814 DASEL-----AQAVKSPFK------VLLKRKYRPQ-----LIIGALGTPAFQQLTGNNSI 945
Query: 298 MLYSPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIG 357
+ Y+P IF+ +G S L + I G +V VI++FL+DK G R+ + G +I
Sbjct: 946 LFYAPVIFQSSGFGSNAALFSSFITNG-ALLVATVISMFLVDKFGTRKFF-LEAGFEMIC 1119
Query: 358 LTLLGLSLTVVDKSNGNVLWALILS--IVATYAYVAFFNIGLGPITWVYSSEIFPLKLRA 415
++ + V+ +G L I + ++ + +V + GP+ W+ SE+FPL++R+
Sbjct: 1120CMIITAVVLAVEFGHGKELSKGISAFLVIMIFWFVLAYGRSWGPLGWLVPSELFPLEIRS 1299
Query: 416 QGASIGVAVNRTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKAL 475
SI V VN A+V+ F+ + G F +F G+ V+ +F +F LPETK +
Sbjct: 1300AAQSIAVCVNMIFTALVAQLFLLSLCHLKY-GIFLLFGGLIVVMSVFVFFLLPETKQVPI 1476
Query: 476 EEMEMVFTKKSSGKNVAIE 494
EE+ ++F KN+ E
Sbjct: 1477EEIYLLFENHWFWKNIVRE 1533
>TC86605 similar to PIR|T01853|T01853 probable hexose transport protein
F9D12.17 - Arabidopsis thaliana, partial (91%)
Length = 3111
Score = 110 bits (276), Expect(2) = 3e-46
Identities = 72/240 (30%), Positives = 123/240 (51%), Gaps = 5/240 (2%)
Frame = +3
Query: 244 ENCNDETVKLPQKSHQGEGV---WKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLY 300
+N E ++L + S + V ++ L+ R R L+ ++ + F+ TGI A+M Y
Sbjct: 2034 DNIEPEFLELVEASRVAKEVKHPFRNLLKRNN---RPQLVISIALMIFQQFTGINAIMFY 2204
Query: 301 SPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTL 360
+P +F G + L A I G ++ +++++ +DKLGRR+LL + M++ +
Sbjct: 2205 APVLFNTLGFKNDAALYSAVI-TGAINVISTIVSIYSVDKLGRRKLLLEAGVQMLLSQMV 2381
Query: 361 LGLSLTVVDKSNGNVLWA--LILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGA 418
+ + L + K + L L +V +V+ F GP+ W+ SEIFPL+ R+ G
Sbjct: 2382 IAIVLGIKVKDHSEELSKGYAALVVVMVCIFVSAFAWSWGPLAWLIPSEIFPLETRSAGQ 2561
Query: 419 SIGVAVNRTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEM 478
S+ V VN AV++ F+S+ G FF F+G + F +F +PETK +EEM
Sbjct: 2562 SVTVCVNFLFTAVIAQAFLSMLCYFKF-GIFFFFSGWILFMSTFVFFLVPETKNVPIEEM 2738
Score = 93.2 bits (230), Expect(2) = 3e-46
Identities = 64/214 (29%), Positives = 102/214 (46%), Gaps = 21/214 (9%)
Frame = +2
Query: 24 IVASMVSIVSGYDTGVMSGAML---FIKEDLGI-------SDTQQ-----------EVLA 62
I+A+ ++ GYD GV G F+K+ SD + ++
Sbjct: 1367 IMAATGGLMFGYDVGVSGGVASMPPFLKKFFPTVLRQTTESDGSESNYCKYDNQGLQLFT 1546
Query: 63 GILNLCALVGSLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVG 122
L L L + A T+ +GRR T+ +A FI G L N +L+VGR + G G
Sbjct: 1547 SSLYLAGLTVTFFASYTTRVLGRRLTMLIAGFFFIAGVSLNASAQNLLMLIVGRVLLGCG 1726
Query: 123 VGFALMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLML 182
+GFA PV+ +EI+ + RG L L +L I +GIL + NY K + GWR+ L
Sbjct: 1727 IGFANQAVPVFLSEIAPSRIRGALNILFQLDITLGILYANLVNYATNK-IKGHWGWRISL 1903
Query: 183 GIAALPSFVVAFCILTMPESPRWLVMQGQLGKAK 216
G+ +P+ ++ + ++P L+ +G LG+ K
Sbjct: 1904 GLGGIPALLLTLGAYLVVDTPNSLIERGHLGQRK 2005
>BF519718 weakly similar to PIR|G84564|G84 probable sugar transporter
[imported] - Arabidopsis thaliana, partial (20%)
Length = 659
Score = 180 bits (456), Expect = 1e-45
Identities = 85/165 (51%), Positives = 123/165 (74%)
Frame = +3
Query: 323 VGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNVLWALILS 382
+G+ K F++ + +LD+ GRR +L + + GM + L LG+ T++ S+ +WA+ L
Sbjct: 9 MGIAKTCFVLFSALVLDRFGRRPMLLLGSSGMAVSLFGLGMGCTLLHNSDEKPMWAIALC 188
Query: 383 IVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTFISIYKA 442
+VA A V+FF+IGLGP TWVYSSEIFP++LRAQG S+ ++VNR ++ VVSM+F+SI +
Sbjct: 189 VVAVCAAVSFFSIGLGPTTWVYSSEIFPMRLRAQGTSLAISVNRLISGVVSMSFLSISEE 368
Query: 443 ITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFTKKSS 487
IT GG FF+ AG+ VLA LFFY+FLPETKGK+LEE+E +F ++ S
Sbjct: 369 ITFGGMFFVLAGVMVLATLFFYYFLPETKGKSLEEIEALFEEELS 503
>TC81556 similar to GP|2688830|gb|AAB88879.1| putative sugar transporter
{Prunus armeniaca}, partial (59%)
Length = 1074
Score = 147 bits (371), Expect = 9e-36
Identities = 103/329 (31%), Positives = 168/329 (50%), Gaps = 7/329 (2%)
Frame = +1
Query: 178 WRLMLGIAALPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIK 237
WR M GIA +PS ++A + PESPRWL QG++ +A+K + + + A + + D++
Sbjct: 19 WRTMFGIAVVPSILLALGMAISPESPRWLFQQGKIAEAEKAIKTLYGKERVATV-MYDLR 195
Query: 238 IAAGLDENCNDETVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVG--IHFFEHATGIE 295
A+ Q S + E W +L S R+ + +VG + F+ GI
Sbjct: 196 TAS--------------QGSSEPEAGWFDLF-----SSRYWKVVSVGAALFLFQQLAGIN 318
Query: 296 AVMLYSPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMI 355
AV+ YS +FR AGI S + A+ VG + ++ IA L+DK GR+ LL S GM
Sbjct: 319 AVVYYSTSVFRSAGIASD---VAASALVGASNVIGTAIASSLMDKQGRKSLLITSFSGMA 489
Query: 356 IGLTLLGLSLT--VVDKSNGNVLWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKL 413
+ LL LS T V+ +G L+++ T YV F++G GP+ + EIF ++
Sbjct: 490 ASMLLLSLSFTWKVLAPYSGT------LAVLGTVLYVLSFSLGAGPVPALLLPEIFASRI 651
Query: 414 RAQGASIGVAVNRTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGK 473
RA+ S+ + + N V+ + F+S+ I + F+ + VLA L+ + ETKG+
Sbjct: 652 RAKAVSLSLGTHWISNFVIGLYFLSVVNKFGISSVYLGFSAVCVLAVLYIAGNVVETKGR 831
Query: 474 ALEEMEMVFTK---KSSGKNVAIEMDPIQ 499
+LEE+E + ++S N + +D Q
Sbjct: 832 SLEEIERALSSSA*ETSRSNCSRNIDASQ 918
>TC79361 similar to GP|17380890|gb|AAL36257.1 putative membrane transporter
protein {Arabidopsis thaliana}, partial (55%)
Length = 949
Score = 140 bits (354), Expect = 9e-34
Identities = 79/210 (37%), Positives = 123/210 (57%), Gaps = 3/210 (1%)
Frame = +1
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKED---LGISDTQQEVLAGILNLCALVG 72
N Y A VA + ++ GYDTGV+SGA+L+IK+D + S+ QE + + A+VG
Sbjct: 160 NPYILGLAAVAGIGGLLFGYDTGVISGALLYIKDDFPQVRNSNFLQETIVSMAIAGAIVG 339
Query: 73 SLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPV 132
+ G +D GR+ LA ++FILGA LM P+ +L+ GR + G+GVG A + APV
Sbjct: 340 AAFGGWLNDAYGRKKATLLADVIFILGAILMAAAPDPYVLIAGRLLVGLGVGIASVTAPV 519
Query: 133 YSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVV 192
Y AE++ + RG L S L I G + Y+ N V + + WR MLG++ +P+ +
Sbjct: 520 YIAEVAPSEIRGSLVSTNVLMITGGQFVSYLVNLV---FTQVPGTWRWMLGVSGVPALIQ 690
Query: 193 AFCILTMPESPRWLVMQGQLGKAKKVLMQV 222
C+L +PESPRWL ++ + +A V+ ++
Sbjct: 691 FICMLFLPESPRWLFIKNRKNEAVDVISKI 780
>TC77798 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_140
{Arabidopsis thaliana}, partial (88%)
Length = 2467
Score = 138 bits (347), Expect = 6e-33
Identities = 73/207 (35%), Positives = 127/207 (61%), Gaps = 1/207 (0%)
Frame = +3
Query: 17 KYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLTA 76
K A AI AS+ + + G+D ++G++L+IK+DL + T + ++ + + A V + +
Sbjct: 114 KGAVLVAIAASIGNFLQGWDNATIAGSILYIKKDLALQTTMEGLVVAMSLIGATVITTCS 293
Query: 77 GRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSAE 136
G SD++GRR + ++S+L+ LG+ +M + PN +L + R + G G+G A+ + PVY +E
Sbjct: 294 GPISDWLGRRPMMIISSVLYFLGSLVMLWSPNVYVLCLARLLDGFGIGLAVTLVPVYISE 473
Query: 137 ISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPS-FVVAFC 195
+ + RG L +LP+ G+ L Y +V+ LS WR+MLG+ ++PS F
Sbjct: 474 TAPSDIRGSLNTLPQFSGSGGMFLSYCMVFVMS--LSPSPSWRIMLGVLSIPSLFYFLLT 647
Query: 196 ILTMPESPRWLVMQGQLGKAKKVLMQV 222
+ +PESPRWLV +G++ +AKKVL ++
Sbjct: 648 VFFLPESPRWLVSKGKMLEAKKVLQRL 728
Score = 109 bits (273), Expect = 2e-24
Identities = 75/243 (30%), Positives = 127/243 (51%), Gaps = 13/243 (5%)
Frame = +3
Query: 254 PQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSK 313
P K+ +W+ L+ P V+ LI +GI + +GI V+ Y+P+I +AG+
Sbjct: 1569 PSKTASKGPIWEALL---EPGVKHALIVGIGIQLLQQFSGINGVLYYTPQILEEAGVA-- 1733
Query: 314 EKLLLATIGVGLTKIVFLV-------------IALFLLDKLGRRRLLQISTGGMIIGLTL 360
+LLA +G+ T FL+ +A+ L+D GRR+LL ++ +I+ L +
Sbjct: 1734 --VLLADLGLSSTSSSFLISAVTTLLMLPSIGLAMRLMDVTGRRQLLLVTIPVLIVSLVI 1907
Query: 361 LGLSLTVVDKSNGNVLWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASI 420
L L +V+D G+V+ A I S V Y FF +G GPI + SEIFP ++R +I
Sbjct: 1908 LVLG-SVIDF--GSVVHAAI-STVCVVVYFCFFVMGYGPIPNILCSEIFPTRVRGLCIAI 2075
Query: 421 GVAVNRTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEM 480
V + +V+ + + ++ + G F ++A + ++W+F Y +PETKG LE +
Sbjct: 2076 CALVFWIGDIIVTYSLPVMLSSLGLAGVFGVYAIVCCISWVFVYLKVPETKGMPLEVITE 2255
Query: 481 VFT 483
F+
Sbjct: 2256 FFS 2264
Score = 31.6 bits (70), Expect = 0.74
Identities = 48/239 (20%), Positives = 103/239 (43%), Gaps = 15/239 (6%)
Frame = +3
Query: 258 HQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGI---TSKE 314
H + ++ L T +++ ++ A+ G + + ++ K + T+ E
Sbjct: 60 HSTQSIYLYFQL*QTQNMKGAVLVAIAASIGNFLQGWDNATIAGSILYIKKDLALQTTME 239
Query: 315 KLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGN 374
L++A +G T V + + D LGRR ++ IS+ +G ++
Sbjct: 240 GLVVAMSLIGAT--VITTCSGPISDWLGRRPMMIISSVLYFLGSLVM------------- 374
Query: 375 VLWALILSIVATYAYVAFFNIGLG-PITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVS 433
LW+ + ++ + F IGL + VY SE P +R S+ + + +
Sbjct: 375 -LWSPNVYVLCLARLLDGFGIGLAVTLVPVYISETAPSDIRG---SLNTLPQFSGSGGMF 542
Query: 434 MTFISIY-KAITIGGSFFMFAGISVLAWLFFY----FFLPE------TKGKALEEMEMV 481
+++ ++ +++ S+ + G+ + LF++ FFLPE +KGK LE +++
Sbjct: 543 LSYCMVFVMSLSPSPSWRIMLGVLSIPSLFYFLLTVFFLPESPRWLVSKGKMLEAKKVL 719
>TC79692 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_140
{Arabidopsis thaliana}, partial (49%)
Length = 1353
Score = 126 bits (317), Expect = 2e-29
Identities = 71/220 (32%), Positives = 126/220 (57%), Gaps = 5/220 (2%)
Frame = +2
Query: 3 INQGDKEDQTNTFNKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLA 62
I+Q KE + A A+ A++ +++ G+D ++G++L+IK + + + +
Sbjct: 128 IDQAKKETMSG-----AVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQS--EPTVE 286
Query: 63 GILNLCALVGSLT----AGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCV 118
G++ +L+G+ +G SD GRR + ++S+L+ L + +M + PN IL+ R +
Sbjct: 287 GLIVAMSLIGATVVTTCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLL 466
Query: 119 CGVGVGFALMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGW 178
G+G+G A+ + P+Y +EI+ RG L +LP+ G+ Y V G L+ W
Sbjct: 467 DGLGIGLAVTLVPLYISEIAPPEIRGSLNTLPQFAGSAGMFFSYC--MVFGMSLTKAPSW 640
Query: 179 RLMLGIAALPSFV-VAFCILTMPESPRWLVMQGQLGKAKK 217
RLMLG+ ++PS + A +L +PESPRWLV +G++ +AKK
Sbjct: 641 RLMLGVLSIPSLIYFALTLLVLPESPRWLVSKGRMLEAKK 760
>TC89747 similar to PIR|F96696|F96696 protein F1N21.12 [imported] -
Arabidopsis thaliana, partial (64%)
Length = 1372
Score = 115 bits (289), Expect = 3e-26
Identities = 98/358 (27%), Positives = 172/358 (47%), Gaps = 4/358 (1%)
Frame = +3
Query: 7 DKEDQTNTFNKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGIS-DTQQEVLAGIL 65
DKE TN K + +VA++ S + GY GV++ + I DLG + +T E L ++
Sbjct: 339 DKET-TNPSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGL--VV 509
Query: 66 NLC---ALVGSLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVG 122
++C AL G L +G +D +GRR L ++ I+GA + N ++VGR G G
Sbjct: 510 SICLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAMSAATNNLFGMLVGRLFVGTG 689
Query: 123 VGFALMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLML 182
+G +A +Y E+S A RG +L ++ GIL S ++ + WR+
Sbjct: 690 LGLGPPVAALYVTEVSPAFVRGTYGALIQIATCFGIL---GSLFIGIPVKEISGWWRVCF 860
Query: 183 GIAALPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGL 242
++ +P+ ++A + ESP WL QG+ +A+ ++ + EA+ + + + +
Sbjct: 861 WVSTIPAAILALAMDFCAESPHWLYKQGRTAEAEAEFERLLGVS-EAKFAMSQL---SKV 1028
Query: 243 DENCNDETVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSP 302
D + +TVK + H K + + T + + +GI AV +S
Sbjct: 1029DRGEDTDTVKFSELLHGHHS--KVVFIGST------------LFALQQLSGINAVFYFSS 1166
Query: 303 RIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTL 360
+F+ AG+ S A + +G+ + +I+ L+DKLGR+ LL S GM I + +
Sbjct: 1167TVFKSAGVPSD----FANVCIGVANLTGSIISTGLMDKLGRKVLLFWSFFGMAISMII 1328
>TC84553 similar to GP|17381265|gb|AAL36051.1 AT5g18840/F17K4_90
{Arabidopsis thaliana}, partial (41%)
Length = 827
Score = 112 bits (281), Expect = 3e-25
Identities = 68/203 (33%), Positives = 111/203 (54%), Gaps = 2/203 (0%)
Frame = +2
Query: 279 LIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFLL 338
++ VG+ + + GI + Y+ F A S + + TI ++ F ++ L+
Sbjct: 8 VVLGVGLMVCQQSVGIMGIGFYTSETFVAADFLSGK---IGTIAYACMQVPFTILGAILM 178
Query: 339 DKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNVLWAL--ILSIVATYAYVAFFNIG 396
DK GRR L+ S G +G + G++ + D+ N+L L IL++ YVA F+IG
Sbjct: 179 DKSGRRPLITASASGTFLGCFMTGIAFFLKDQ---NLLLELVPILAVAGILIYVAAFSIG 349
Query: 397 LGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTFISIYKAITIGGSFFMFAGIS 456
+GP+ WV SEIFP+ ++ S+ V +N VVS TF + + + + G+ F++AG S
Sbjct: 350 MGPVPWVIMSEIFPIHVKGTAGSLVVLINWLGAWVVSYTF-NFFLSWSSPGTLFLYAGCS 526
Query: 457 VLAWLFFYFFLPETKGKALEEME 479
+L LF +PETKGK LEE++
Sbjct: 527 LLTILFVAKLVPETKGKTLEEIQ 595
>TC89865 similar to GP|17380890|gb|AAL36257.1 putative membrane transporter
protein {Arabidopsis thaliana}, partial (33%)
Length = 617
Score = 105 bits (261), Expect = 5e-23
Identities = 60/160 (37%), Positives = 94/160 (58%), Gaps = 3/160 (1%)
Frame = +1
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKED---LGISDTQQEVLAGILNLCALVG 72
N Y VAS+ ++ GYDTGV+SGA+L+IK+D + S QE + + A+VG
Sbjct: 82 NPYILCLTSVASIGGLLFGYDTGVISGALLYIKDDFQAVRYSHFLQETIVSMAVAGAIVG 261
Query: 73 SLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPV 132
+ G +D GR+ +A ++FILGA +M P+ IL++GR + G+GVG A + APV
Sbjct: 262 AAVGGWMNDRYGRKKATIIADVIFILGAIVMAAAPDPYILILGRVLVGLGVGIASVTAPV 441
Query: 133 YSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYL 172
Y AE+S + RG L + L I G + Y+ ++L +++
Sbjct: 442 YIAELSPSEIRGGLVATNVLMITGGQFISYLVIFLLRRFI 561
>TC84386 similar to GP|21689841|gb|AAM67564.1 unknown protein {Arabidopsis
thaliana}, partial (30%)
Length = 664
Score = 100 bits (250), Expect = 1e-21
Identities = 60/153 (39%), Positives = 82/153 (53%), Gaps = 3/153 (1%)
Frame = +2
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQ---QEVLAGILNLCALVG 72
N Y A A + + GYDTGV+SGA+L+I++D D Q QE + A++G
Sbjct: 176 NPYVLRLAFSAGIGGFLFGYDTGVISGALLYIRDDFKAVDRQTWLQEAIVSTALAGAIIG 355
Query: 73 SLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPV 132
+ G +D GR+ I LA LF +G+ +M N AIL+VGR G+GVG A M +P+
Sbjct: 356 ASVGGWINDRFGRKKAIILADALFFIGSVIMAAAINPAILIVGRVFVGLGVGMASMASPL 535
Query: 133 YSAEISSASSRGFLTSLPELCIGIGILLGYISN 165
Y +E S RG L SL I G L Y+ N
Sbjct: 536 YISEASPTRVRGALVSLNGFLITGGQXLSYVIN 634
>TC81269 similar to GP|23504385|emb|CAC00697. putative sugar transporter
{Lycopersicon esculentum}, partial (42%)
Length = 966
Score = 100 bits (248), Expect = 2e-21
Identities = 68/203 (33%), Positives = 104/203 (50%), Gaps = 2/203 (0%)
Frame = +2
Query: 279 LIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFLL 338
L VG+ + GI V Y+ IF AG S ++ I +IV + L+
Sbjct: 41 LTIGVGLMVCQQLGGINGVCFYTSSIFDLAGFPSATGSIIYAI----LQIVITGVGAALI 208
Query: 339 DKLGRRRLLQISTGGMIIG--LTLLGLSLTVVDKSNGNVLWALILSIVATYAYVAFFNIG 396
D+ GR+ LL +S G++ G T + L V D + G V L++ Y+ F+IG
Sbjct: 209 DRAGRKPLLLVSGSGLVAGCIFTAVAFYLKVHDVAVGAVP---ALAVTGILVYIGSFSIG 379
Query: 397 LGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTFISIYKAITIGGSFFMFAGIS 456
+G I WV SEIFP+ ++ Q SI VN + S TF + + + G+F ++A I+
Sbjct: 380 MGAIPWVVMSEIFPVNIKGQAGSIATLVNWFGAWLCSYTF-NFLMSWSSYGTFVLYAAIN 556
Query: 457 VLAWLFFYFFLPETKGKALEEME 479
LA LF +PETKGK+LE+++
Sbjct: 557 ALAILFIAVVVPETKGKSLEQLQ 625
>BQ751130 weakly similar to GP|15289796|dbj putative monosaccharide
transporter 3 {Oryza sativa (japonica cultivar-group)},
partial (7%)
Length = 795
Score = 99.8 bits (247), Expect = 2e-21
Identities = 69/232 (29%), Positives = 115/232 (48%), Gaps = 20/232 (8%)
Frame = +3
Query: 19 AFACAIVASMVSIVSGYDTGVMSGAM---LFIKEDLG-----ISDTQQEVLAGILNLCAL 70
A+ AS I GYD+G ++G + FI+ G IS++ Q ++ IL+
Sbjct: 39 AYVICAFASFGGIFFGYDSGYINGVLGANQFIEHVEGPGAEAISESHQSLIVSILSCGTF 218
Query: 71 VGSLTAGRTSDYIGRRYTIFLASILFILGAGLM---GYGPNYAILMVGRCVCGVGVGFAL 127
G+L AG SD+IGR++T+ + ++++G + G G ++ GR + G+GVGF
Sbjct: 219 FGALIAGDVSDWIGRKWTVIIGCGIYMIGVLIQMFTGMGAPLGAIVAGRLIAGIGVGFES 398
Query: 128 MIAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYV------LGKY---LSLKLGW 178
I +Y +EI RG L + + CI IG+LL Y G Y ++++ W
Sbjct: 399 AIVILYMSEICPKKVRGALVAGYQFCITIGLLLAACVVYATKDRGDTGVYRIPIAIQFPW 578
Query: 179 RLMLGIAALPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAE 230
L+LG +L +PESPR+ V +G++ A L ++ +E+E
Sbjct: 579 ALILGGG----------LLLLPESPRFFVKKGRIADATAALSRLRGQPKESE 704
>TC86673 weakly similar to PIR|S25015|S25015 monosaccharide transport
protein MST1 - common tobacco, partial (65%)
Length = 1162
Score = 98.6 bits (244), Expect = 5e-21
Identities = 70/215 (32%), Positives = 103/215 (47%), Gaps = 21/215 (9%)
Frame = +2
Query: 24 IVASMVSIVSGYDTGVMSGAML---FIKEDLGISDTQQEVLAGILN-------------- 66
I+A+ ++ GYD GV G F+K Q+ L N
Sbjct: 110 IMAATGGLIFGYDHGVSGGVTSMDSFLKRFFPSVYEQESNLKPSANQYCKFNSQILTLFT 289
Query: 67 ----LCALVGSLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVG 122
+ ALV L A + +GRR T+ L I F+ GA L G N A+L+ GR + G G
Sbjct: 290 SSLYISALVAGLGASSITRALGRRTTMILGGIFFVSGALLNGLAMNIAMLIAGRLLLGFG 469
Query: 123 VGFALMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLML 182
+G A P+Y +E++ RG L +L I IGI + NY K L+ + GWRL L
Sbjct: 470 IGCANQAVPIYLSEMAPYKYRGALNMCFQLSITIGIFTANMFNYYFSKILNGE-GWRLSL 646
Query: 183 GIAALPSFVVAFCILTMPESPRWLVMQGQLGKAKK 217
G+ A+P+ V + +P+SP LV +G+ +A+K
Sbjct: 647 GLGAVPAVVFIIGSICLPDSPNSLVTRGRHEEARK 751
Score = 55.5 bits (132), Expect = 5e-08
Identities = 34/104 (32%), Positives = 56/104 (53%)
Frame = +3
Query: 264 WKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKLLLATIGV 323
WK L R R L+ A+ I FF+ TG+ + Y+P +FR G S+ L+ A I +
Sbjct: 846 WKTLQER---KYRPQLVFAILIPFFQQFTGLNVITFYAPILFRTIGFGSQASLMSAAI-I 1013
Query: 324 GLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTV 367
G K V ++++F++DK GRR L M+I ++ +++ V
Sbjct: 1014GSFKPVSTLVSIFVVDKFGRRALFLEGGVQMLISXIIMTVAIAV 1145
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.141 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,442,475
Number of Sequences: 36976
Number of extensions: 200480
Number of successful extensions: 1579
Number of sequences better than 10.0: 108
Number of HSP's better than 10.0 without gapping: 1474
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1507
length of query: 501
length of database: 9,014,727
effective HSP length: 100
effective length of query: 401
effective length of database: 5,317,127
effective search space: 2132167927
effective search space used: 2132167927
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147964.1


