

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147963.9 - phase: 0
(423 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
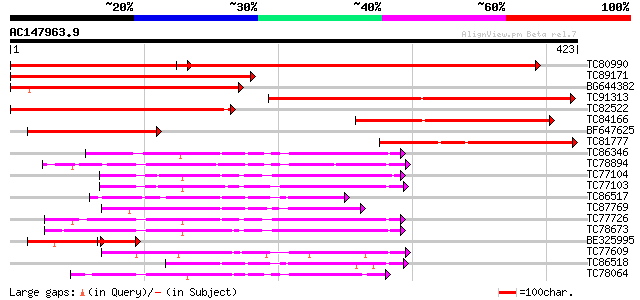
Score E
Sequences producing significant alignments: (bits) Value
TC80990 similar to PIR|A86224|A86224 hypothetical protein [impor... 513 e-146
TC89171 similar to GP|16930415|gb|AAL31893.1 At1g09160/T12M4_13 ... 320 7e-88
BG644382 similar to GP|16930415|gb At1g09160/T12M4_13 {Arabidops... 278 3e-75
TC91313 similar to GP|15292869|gb|AAK92805.1 unknown protein {Ar... 241 3e-64
TC82522 similar to PIR|A96708|A96708 hypothetical protein T2E12.... 226 1e-59
TC84166 similar to PIR|A96708|A96708 hypothetical protein T2E12.... 156 1e-38
BF647625 similar to GP|15292869|gb unknown protein {Arabidopsis ... 120 8e-28
TC81777 similar to GP|15292869|gb|AAK92805.1 unknown protein {Ar... 117 8e-27
TC86346 homologue to GP|22136752|gb|AAM91695.1 unknown protein {... 117 1e-26
TC78894 similar to GP|17381034|gb|AAL36329.1 putative protein ph... 109 2e-24
TC77104 similar to GP|4336436|gb|AAD17805.1| protein phosphatase... 94 8e-20
TC77103 similar to GP|4336436|gb|AAD17805.1| protein phosphatase... 91 1e-18
TC86517 homologue to PIR|T09640|T09640 protein phosphatase 2C - ... 90 1e-18
TC87769 similar to GP|14334800|gb|AAK59578.1 putative protein ph... 89 3e-18
TC77726 similar to PIR|F86355|F86355 hypothetical protein AAF872... 87 2e-17
TC78673 similar to GP|13877699|gb|AAK43927.1 protein phosphatase... 85 5e-17
BE325995 similar to GP|15292869|gb| unknown protein {Arabidopsis... 52 2e-16
TC77609 weakly similar to GP|9909177|dbj|BAB12036.1 putative pro... 83 2e-16
TC86518 homologue to PIR|T09640|T09640 protein phosphatase 2C - ... 83 2e-16
TC78064 similar to GP|20259167|gb|AAM14299.1 putative phosphatas... 82 5e-16
>TC80990 similar to PIR|A86224|A86224 hypothetical protein [imported] -
Arabidopsis thaliana, partial (71%)
Length = 1343
Score = 513 bits (1321), Expect = e-146
Identities = 256/272 (94%), Positives = 259/272 (95%)
Frame = +3
Query: 125 QKKGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVT 184
+K+G FVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVT
Sbjct: 513 EKRGNLLERQLPFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVT 692
Query: 185 ASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRL 244
ASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRL
Sbjct: 693 ASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRL 872
Query: 245 IIASDGIWDTLSSDMAAKSCRGVPAELAAKLVVKEALRSRGLKDDTTCLVVDIIPSDYPV 304
IIASDGIWDTLSSDMAAKSCRGVPAELAAKLVVKEALRSRGLKDDTTCLVVDIIPSDYPV
Sbjct: 873 IIASDGIWDTLSSDMAAKSCRGVPAELAAKLVVKEALRSRGLKDDTTCLVVDIIPSDYPV 1052
Query: 305 LPMPATPRKKHNVLSSLLFGKKSQNLANKGTNKLSAVGVVEELFEEGSAMLTERLGNNVP 364
LPMPATP+KKHNVLSSLLFGKKSQNLANKGTNKLSAVGVVEELFEEGSAMLTE LGNNVP
Sbjct: 1053LPMPATPKKKHNVLSSLLFGKKSQNLANKGTNKLSAVGVVEELFEEGSAMLTEXLGNNVP 1232
Query: 365 SDTNSGIHRCAVCLADQPSGDGLSVNNDHFIT 396
DTNSGIHRCAVCL QPSGDGLS NNDHFIT
Sbjct: 1233XDTNSGIHRCAVCLTXQPSGDGLSXNNDHFIT 1328
Score = 196 bits (497), Expect = 2e-50
Identities = 107/137 (78%), Positives = 113/137 (82%), Gaps = 1/137 (0%)
Frame = +2
Query: 1 MSKAELSRMKQPLVPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPG 60
MSKAELSRMKQPLVPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPG
Sbjct: 137 MSKAELSRMKQPLVPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPG 316
Query: 61 DSSTSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFV-K 119
DSSTSFSVFAILDGHNGISAAIFAKENIINNVMS + L L ++ + K
Sbjct: 317 DSSTSFSVFAILDGHNGISAAIFAKENIINNVMSCHTARL*AGRNGFKLCLGL*LSLL*K 496
Query: 120 TDMEFQKKGETSGTTAT 136
F+KKG+ SGTTAT
Sbjct: 497 LTWNFRKKGKPSGTTAT 547
>TC89171 similar to GP|16930415|gb|AAL31893.1 At1g09160/T12M4_13
{Arabidopsis thaliana}, partial (49%)
Length = 729
Score = 320 bits (820), Expect = 7e-88
Identities = 158/183 (86%), Positives = 170/183 (92%)
Frame = +2
Query: 1 MSKAELSRMKQPLVPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPG 60
MSK ELSRMK VPLATLIGRE+RN K EKPFVK+GQAGLAKKGEDYFLIKTDCHRVPG
Sbjct: 179 MSKRELSRMKPSPVPLATLIGREIRNEKVEKPFVKFGQAGLAKKGEDYFLIKTDCHRVPG 358
Query: 61 DSSTSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKT 120
D ST+FSVFAI DGHNGISAAI+ KEN++NNV+SAIPQ +SR+ WLQALPRALVV FVKT
Sbjct: 359 DPSTAFSVFAIFDGHNGISAAIYTKENLLNNVLSAIPQDISRDAWLQALPRALVVGFVKT 538
Query: 121 DMEFQKKGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEER 180
D EFQKKGETSGTTATFVI++GWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENV+ER
Sbjct: 539 DTEFQKKGETSGTTATFVIVEGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVKER 718
Query: 181 ERV 183
ERV
Sbjct: 719 ERV 727
>BG644382 similar to GP|16930415|gb At1g09160/T12M4_13 {Arabidopsis
thaliana}, partial (45%)
Length = 701
Score = 278 bits (711), Expect = 3e-75
Identities = 139/176 (78%), Positives = 155/176 (87%), Gaps = 2/176 (1%)
Frame = +3
Query: 1 MSKAELSRMKQPL--VPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRV 58
MS+AE++RMKQ VPL TLIGRELRN K E+ +KYGQA LAKKGED FLIK DC R+
Sbjct: 174 MSEAEITRMKQKTTSVPLGTLIGRELRNDKVEQRTIKYGQAALAKKGEDCFLIKLDCQRI 353
Query: 59 PGDSSTSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFV 118
PG+ TSFSVFAI DGHNGISAAIFAKEN++NNV+SAIP+G+SREEWLQAL RALV FV
Sbjct: 354 PGNPLTSFSVFAIFDGHNGISAAIFAKENLLNNVLSAIPEGISREEWLQALRRALVAGFV 533
Query: 119 KTDMEFQKKGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLE 174
KTD+EFQ+KGETSGTT TFV+IDGWT+TVASVGDSRCILDTQGGVVSLLT DHRLE
Sbjct: 534 KTDIEFQQKGETSGTTVTFVVIDGWTITVASVGDSRCILDTQGGVVSLLTGDHRLE 701
>TC91313 similar to GP|15292869|gb|AAK92805.1 unknown protein {Arabidopsis
thaliana}, partial (54%)
Length = 806
Score = 241 bits (616), Expect = 3e-64
Identities = 119/229 (51%), Positives = 159/229 (68%)
Frame = +1
Query: 194 NVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWD 253
N GG EVGPLRCWPGGLCLSRSIGD DVGE+IVP+P+VKQVKLSNAGGRL+I+SDG+WD
Sbjct: 22 NTGGGAEVGPLRCWPGGLCLSRSIGDMDVGEFIVPVPYVKQVKLSNAGGRLVISSDGVWD 201
Query: 254 TLSSDMAAKSCRGVPAELAAKLVVKEALRSRGLKDDTTCLVVDIIPSDYPVLPMPATPRK 313
L+++M SCRG+ AE AA +VKE+L+++GL+DDTTC+VVDI+P + P + T +K
Sbjct: 202 ALTAEMVLDSCRGMSAEAAAPHIVKESLQAKGLRDDTTCVVVDILPQEKPPTSV-RTQKK 378
Query: 314 KHNVLSSLLFGKKSQNLANKGTNKLSAVGVVEELFEEGSAMLTERLGNNVPSDTNSGIHR 373
+ +F KKS ++ + +V EL+EEGSA+L+ER P +
Sbjct: 379 PVKGMLKAIFRKKSSESSSYVEKEYVEPDMVRELYEEGSALLSERFETKYPLCNMFKLFM 558
Query: 374 CAVCLADQPSGDGLSVNNDHFITPVSKPWEGPFLCTNCQKKKDAMEGKR 422
CAVC + G+G+SV+ +PW+GPFLC +CQKKK+AME K+
Sbjct: 559 CAVCQVEIKPGEGISVHEGAPNARKPRPWDGPFLCLSCQKKKEAMERKQ 705
>TC82522 similar to PIR|A96708|A96708 hypothetical protein T2E12.9
[imported] - Arabidopsis thaliana, partial (33%)
Length = 727
Score = 226 bits (576), Expect = 1e-59
Identities = 112/168 (66%), Positives = 136/168 (80%)
Frame = +2
Query: 1 MSKAELSRMKQPLVPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPG 60
M+ E R LVPLA L+ RE+++ K EKP V+ GQA +KKGEDYFLIK DC RVPG
Sbjct: 227 MASGEGRRHHHDLVPLAALLKREMKSEKMEKPTVRIGQAAQSKKGEDYFLIKIDCQRVPG 406
Query: 61 DSSTSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKT 120
+SSTSFSVFAI DGHNG +AA++ KEN++N+V+ A+P+G+ R+EWLQALPRALV FVKT
Sbjct: 407 NSSTSFSVFAIFDGHNGNAAAVYTKENLLNHVLCALPRGLGRDEWLQALPRALVAGFVKT 586
Query: 121 DMEFQKKGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLT 168
D EFQ +GETSGTTATFVI+D WTVTVASVGDSRCILD + VV++ T
Sbjct: 587 DKEFQSRGETSGTTATFVIVDRWTVTVASVGDSRCILDPR--VVAVTT 724
>TC84166 similar to PIR|A96708|A96708 hypothetical protein T2E12.9
[imported] - Arabidopsis thaliana, partial (34%)
Length = 878
Score = 156 bits (395), Expect = 1e-38
Identities = 83/149 (55%), Positives = 106/149 (70%), Gaps = 1/149 (0%)
Frame = +3
Query: 259 MAAKSCRGVPAELAAKLVVKEALRSRGLKDDTTCLVVDIIPSDYPVLPMPATPRKKHNVL 318
+AAKSCRG+PAELAA VVKEALR+RGLKDDTTC+VVDIIP D + P P P+K++ +
Sbjct: 3 LAAKSCRGLPAELAAMQVVKEALRTRGLKDDTTCIVVDIIPPDNELPPTP-PPKKRNKLK 179
Query: 319 SSLLFGKKSQNLANKGTNKLSAVGVVEELFEEGSAMLTERLGNNVPS-DTNSGIHRCAVC 377
F K+S++ A K + KLSA+ +VEELFEEGSAML ERLGN+ S + SG+ CAVC
Sbjct: 180 DFFSFKKRSRDTAGKLSKKLSAINIVEELFEEGSAMLAERLGNDENSGQSTSGLFVCAVC 359
Query: 378 LADQPSGDGLSVNNDHFITPVSKPWEGPF 406
D +G+SV+ + SKPW+G F
Sbjct: 360 QLDLAPSEGISVHAGSIFSTSSKPWQGSF 446
Score = 38.9 bits (89), Expect = 0.004
Identities = 17/29 (58%), Positives = 20/29 (68%)
Frame = +1
Query: 394 FITPVSKPWEGPFLCTNCQKKKDAMEGKR 422
F V +GPFLC +C+ KKDAMEGKR
Sbjct: 409 FSPQVRSHGKGPFLCCDCRDKKDAMEGKR 495
>BF647625 similar to GP|15292869|gb unknown protein {Arabidopsis thaliana},
partial (24%)
Length = 655
Score = 120 bits (302), Expect = 8e-28
Identities = 55/100 (55%), Positives = 78/100 (78%)
Frame = +1
Query: 14 VPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPGDSSTSFSVFAILD 73
VPL+ L+ REL + K EKP + +GQAG +KKGED+ L+K+DC R+ GD ++SV+ + D
Sbjct: 355 VPLSVLLKRELMSEKIEKPEIVHGQAGQSKKGEDFALLKSDCQRMVGDGVYTYSVYGLFD 534
Query: 74 GHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRAL 113
GHNG +AAI++ EN++NNV+SAIP ++R EWL ALP+AL
Sbjct: 535 GHNGSAAAIYSXENLLNNVVSAIPPDLNRXEWLXALPKAL 654
>TC81777 similar to GP|15292869|gb|AAK92805.1 unknown protein {Arabidopsis
thaliana}, partial (40%)
Length = 951
Score = 117 bits (293), Expect = 8e-27
Identities = 63/147 (42%), Positives = 91/147 (61%)
Frame = +3
Query: 277 VKEALRSRGLKDDTTCLVVDIIPSDYPVLPMPATPRKKHNVLSSLLFGKKSQNLANKGTN 336
++EAL+++GL+DDTTC+VVDI+P + P P+P + +L + +F KKS ++
Sbjct: 294 LQEALQAKGLRDDTTCVVVDILPQEKPPAPVPHQKKPVKGMLKA-MFRKKSSESSSIDKE 470
Query: 337 KLSAVGVVEELFEEGSAMLTERLGNNVPSDTNSGIHRCAVCLADQPSGDGLSVNNDHFIT 396
+ VV EL+EEGSAML+ERL P + CAVC G+G+SV+
Sbjct: 471 YMEP-DVVVELYEEGSAMLSERLKTKYPLCNMFKLFICAVCQVXIKPGEGISVHEGAXNP 647
Query: 397 PVSKPWEGPFLCTNCQKKKDAMEGKRS 423
+PW+GPFLC +C +KK+AMEGKRS
Sbjct: 648 QKVRPWDGPFLCFSCXEKKEAMEGKRS 728
Score = 38.5 bits (88), Expect = 0.005
Identities = 16/31 (51%), Positives = 22/31 (70%)
Frame = +2
Query: 248 SDGIWDTLSSDMAAKSCRGVPAELAAKLVVK 278
SDG+WD L +++A CRG+ AE AA +VK
Sbjct: 5 SDGVWDALPAEVALDCCRGMSAESAAPQIVK 97
>TC86346 homologue to GP|22136752|gb|AAM91695.1 unknown protein {Arabidopsis
thaliana}, partial (89%)
Length = 2436
Score = 117 bits (292), Expect = 1e-26
Identities = 80/245 (32%), Positives = 139/245 (56%), Gaps = 6/245 (2%)
Frame = +1
Query: 57 RVPGDSSTSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVA 116
R+ G + +F + DGH G AA + K+N+ +N+ +S +++ A+ A
Sbjct: 481 RIDGINGEVVGLFGVFDGHGGARAAEYVKQNLFSNL-------ISHPKFISDTKSAIADA 639
Query: 117 FVKTDMEFQK----KGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHR 172
+ TD EF K + +G+TA+ I+ G + VA+VGDSR ++ +GG ++ DH+
Sbjct: 640 YTHTDSEFLKSENNQNRDAGSTASTAILVGDRLLVANVGDSRAVI-CRGGNAIAVSRDHK 816
Query: 173 LEENVEERERVTASGGEVGRLNVFGGN-EVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPH 231
++ +ER+R+ +GG V ++ G VG G L +SR+ GD + +Y+V P
Sbjct: 817 PDQT-DERQRIEDAGGFV----MWAGTWRVG------GVLAVSRAFGDRLLKQYVVADPE 963
Query: 232 VKQVKLSNAGGRLIIASDGIWDTLSSDMAAKSCRGV-PAELAAKLVVKEALRSRGLKDDT 290
+++ K+ ++ LI+ASDG+WD +S++ A + + AE AAK ++KEA + RG D+
Sbjct: 964 IQEEKVDSSLEFLILASDGLWDVVSNEEAVAMIKPIEDAEEAAKRLMKEAYQ-RGSSDNI 1140
Query: 291 TCLVV 295
TC+VV
Sbjct: 1141TCVVV 1155
>TC78894 similar to GP|17381034|gb|AAL36329.1 putative protein phosphatase
type 2C {Arabidopsis thaliana}, partial (71%)
Length = 1227
Score = 109 bits (272), Expect = 2e-24
Identities = 89/281 (31%), Positives = 150/281 (52%), Gaps = 6/281 (2%)
Frame = +1
Query: 25 RNGKTEKPFVKYGQAGLAKKG---EDYFLIKTDCHRVPGDSSTSFSVFAILDGHNGISAA 81
RNG+ + YG + K ED+F +T V G F VF DGH G A
Sbjct: 229 RNGR-----ISYGYSSFKGKRSSMEDFF--ETKVSEVDGQMVGFFGVF---DGHGGSRTA 378
Query: 82 IFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKKGET-SGTTATFVII 140
+ K N+ N+ S +++ A+V AF +TD+++ K+ +G+TA+ ++
Sbjct: 379 EYLKNNLFKNLCS-------HPDFITDTKTAIVEAFKQTDVDYLKEEMGHAGSTASTAVL 537
Query: 141 DGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEVGRLNVFGGN- 199
G + VA+VGDSR ++ ++ G+ L+VDH+ + +ER+R+ +GG V ++ G
Sbjct: 538 LGDRILVANVGDSR-VVASRAGLAVPLSVDHKPNRS-DERQRIEQAGGFV----IWAGTW 699
Query: 200 EVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSDM 259
VG G L +SR+ GD + Y+V P +++ ++ +IIASDG+WD +S++
Sbjct: 700 RVG------GILAVSRAFGDKLLRPYVVADPEIQEEEIEGVDF-IIIASDGLWDVISNEE 858
Query: 260 AAKSCRG-VPAELAAKLVVKEALRSRGLKDDTTCLVVDIIP 299
A + AE+A++ ++KEA +RG D+ TC+VV P
Sbjct: 859 AVSLVQNFTDAEVASRELIKEAY-TRGSSDNITCVVVRFDP 978
>TC77104 similar to GP|4336436|gb|AAD17805.1| protein phosphatase type 2C
{Lotus japonicus}, complete
Length = 1199
Score = 94.4 bits (233), Expect = 8e-20
Identities = 70/233 (30%), Positives = 118/233 (50%), Gaps = 5/233 (2%)
Frame = +3
Query: 68 VFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKK 127
+FAI DGH G + A + + N+ +N++ + W + + ++ A+ +TD +K
Sbjct: 291 LFAIFDGHAGHTVADYLRSNLFDNILK------EPDFWTEPVD-SVKRAYKETDSTILEK 449
Query: 128 ----GETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERV 183
G+ T T ++I+ + VA++GDSR +L ++ GV L+VDH E E +
Sbjct: 450 SGELGKGGSTAVTAILINCQKLVVANIGDSRAVL-SENGVAIPLSVDH---EPTTESNDI 617
Query: 184 TASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGR 243
GG F N G + G L +SR+ GD + ++ PHV + + G
Sbjct: 618 KNRGG-------FVSNFPGDVPRVDGQLAVSRAFGDKSLKIHMTSEPHVTVKMIDDGGEF 776
Query: 244 LIIASDGIWDTLSSDMAAKSCRGV-PAELAAKLVVKEALRSRGLKDDTTCLVV 295
+I+ASDG+W +S+ A + + + A AAK + +EAL R DD +C+VV
Sbjct: 777 VILASDGLWKVMSNQEAVDAIKDIKDARSAAKHLTEEALNRRS-SDDISCIVV 932
>TC77103 similar to GP|4336436|gb|AAD17805.1| protein phosphatase type 2C
{Lotus japonicus}, partial (98%)
Length = 1241
Score = 90.5 bits (223), Expect = 1e-18
Identities = 69/235 (29%), Positives = 117/235 (49%), Gaps = 5/235 (2%)
Frame = +3
Query: 68 VFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKK 127
+FAI DGH G + + + ++ +N++ + W + + A+ A+ TD +K
Sbjct: 315 LFAIFDGHAGHNVPNYLQSHLFDNILK------EPDFWTEPV-NAVKEAYSITDSSILEK 473
Query: 128 ----GETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERV 183
G T T ++IDG + VA++GDSR +L +G V L+VDH E E E +
Sbjct: 474 SGELGRGGSTAVTAILIDGQKLVVANIGDSRAVLSKKG-VAKQLSVDH---EPTTEHEDI 641
Query: 184 TASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGR 243
GG V R G + G L +SR+ GD + +++ P+V K+S
Sbjct: 642 KNRGGFVSRFP-------GDVPRVDGRLAVSRAFGDKSLKKHLSSEPYVTVEKISEDAEF 800
Query: 244 LIIASDGIWDTLSSDMAAKSCRGV-PAELAAKLVVKEALRSRGLKDDTTCLVVDI 297
+I+ASDG+W +S+ A + + A +AK + +EA+ +R DD + +VV +
Sbjct: 801 VILASDGLWKVMSNQEAVDCIKDIKDARSSAKRLTEEAV-NRKSSDDISVIVVKL 962
>TC86517 homologue to PIR|T09640|T09640 protein phosphatase 2C - alfalfa,
complete
Length = 1616
Score = 90.1 bits (222), Expect = 1e-18
Identities = 59/194 (30%), Positives = 97/194 (49%)
Frame = +2
Query: 60 GDSSTSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVK 119
G+++ +F F + DGH G AA FA N+ N++ + +++ +A+ R ++
Sbjct: 572 GENNLAF--FGVFDGHGGAKAAEFAANNLEKNILDEVIMS-DKDDVEEAVKRG----YLN 730
Query: 120 TDMEFQKKGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEE 179
TD EF KK G+ I + V++ GD R ++ ++GGV LT DHR +E
Sbjct: 731 TDSEFMKKDLHGGSCCVTAFIRNGNLVVSNAGDCRAVI-SRGGVAEALTSDHRPSRE-DE 904
Query: 180 RERVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSN 239
++R+ GG V G R G L +SR IGD + +++ P K +++
Sbjct: 905 KDRIETLGGYVDLCR-------GVWRI-QGSLAVSRGIGDRHLKQWVTAEPETKVIRIEP 1060
Query: 240 AGGRLIIASDGIWD 253
LI+ASDG+WD
Sbjct: 1061EHDLLILASDGLWD 1102
>TC87769 similar to GP|14334800|gb|AAK59578.1 putative protein phosphatase
ABI1 {Arabidopsis thaliana}, partial (56%)
Length = 1137
Score = 89.0 bits (219), Expect = 3e-18
Identities = 65/205 (31%), Positives = 100/205 (48%), Gaps = 8/205 (3%)
Frame = +1
Query: 69 FAILDGHNGISAAIFAKENI-------INNVMSAIPQGVSREEWLQALPRALVVAFVKTD 121
F + DGH G A + +E + I S++ + R W + + L F K D
Sbjct: 190 FGVYDGHGGFQVANYCQERLHSALIEEIEARQSSLDETNGRNNWQENWNKVLFNCFQKVD 369
Query: 122 MEFQKKG-ETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEER 180
E + ET+G+TA I++ + VA+ GDSR +L +G L+ DH+ +ER
Sbjct: 370 DEIETLAPETAGSTAVVAILNQTHIIVANCGDSRAVL-YRGKEAIALSSDHKPNRE-DER 543
Query: 181 ERVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNA 240
R+ A+GG V + G V G L +SRSIGD + +I+P P V V+
Sbjct: 544 ARIEAAGGRVIH---WKGYRV------LGVLAMSRSIGDRYLKPWIIPEPEVNMVQREKN 696
Query: 241 GGRLIIASDGIWDTLSSDMAAKSCR 265
LI+ASDG+WD ++++ A R
Sbjct: 697 DECLILASDGLWDVMTNEEACDIAR 771
>TC77726 similar to PIR|F86355|F86355 hypothetical protein AAF87263.1
[imported] - Arabidopsis thaliana, partial (91%)
Length = 1510
Score = 86.7 bits (213), Expect = 2e-17
Identities = 76/278 (27%), Positives = 131/278 (46%), Gaps = 9/278 (3%)
Frame = +2
Query: 27 GKTEKPFVKYGQAGLAKKG----EDYFLIKTDCHRVPGDSSTSFSVFAILDGHNGISAAI 82
GK+ + VKYG + + K EDY + K + +FAI DGH G S
Sbjct: 413 GKSSQSPVKYGFSLVKGKANHPMEDYHVAK-----IVHFKGQELGLFAIYDGHLGDSVPS 577
Query: 83 FAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKK----GETSGTTATFV 138
+ ++++ +N++ +++ ++ A+ TD G T T +
Sbjct: 578 YLQKHLFSNILK-------EDDFWNDPFMSISKAYESTDQAILTHSPDLGRGGSTAVTAI 736
Query: 139 IIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEVGRLNVFGG 198
+I+ + VA+VGDSR +L ++GGV ++ DH E ER + GG F
Sbjct: 737 LINNQKLWVANVGDSRAVL-SRGGVAVQMSTDH---EPNTERGSIENRGG-------FVS 883
Query: 199 NEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSD 258
N G + G L +SR+ GD ++ ++ P ++ ++ LI+ASDG+W +++
Sbjct: 884 NMPGDVARVNGQLAVSRAFGDKNLKTHLRSDPDIQYADVNQDTEFLILASDGLWKVMANQ 1063
Query: 259 MAAKSCRGV-PAELAAKLVVKEALRSRGLKDDTTCLVV 295
A + + + AAK + EAL +R KDD +C+VV
Sbjct: 1064EAVDIAKKIKDPQKAAKQLATEAL-NRDSKDDISCVVV 1174
>TC78673 similar to GP|13877699|gb|AAK43927.1 protein phosphatase type
2C-like protein {Arabidopsis thaliana}, partial (94%)
Length = 1529
Score = 85.1 bits (209), Expect = 5e-17
Identities = 68/274 (24%), Positives = 131/274 (46%), Gaps = 5/274 (1%)
Frame = +2
Query: 27 GKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPGDSSTSFSVFAILDGHNGISAAIFAKE 86
GK+ + +KYG L + +D+ + ++ + +FAI DGH+G + + ++
Sbjct: 476 GKSLQCPLKYGFC-LVEGTQDHTMEDFHVAKIVQFNGRELGLFAIFDGHSGDTVPAYLQK 652
Query: 87 NIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKK----GETSGTTATFVIIDG 142
++ +N++ E++ +++ A+ TD G T T ++++
Sbjct: 653 HLFSNILK-------EEDFWTDPNSSIIEAYEATDQAILSHSPDLGRGGSTAVTAILVNN 811
Query: 143 WTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEVGRLNVFGGNEVG 202
+ +A+VGDSR +L +G + + ++DH E ER + GG F N G
Sbjct: 812 QKLWIANVGDSRAVLSRKGVAIQM-SIDH---EPNTERRIIENKGG-------FVSNLPG 958
Query: 203 PLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSDMAAK 262
+ G L ++R+ GD ++ ++ P VK + LI+ASDG+W +++ A
Sbjct: 959 DVARVNGQLAVARAFGDRNLKSHLRSDPDVKPDDIDQDTELLILASDGLWKVMANQEAVD 1138
Query: 263 SCRGV-PAELAAKLVVKEALRSRGLKDDTTCLVV 295
+ + AAK ++ EAL+ R +DD +C+VV
Sbjct: 1139IALKIKDPQKAAKQLIAEALK-RESRDDISCIVV 1237
>BE325995 similar to GP|15292869|gb| unknown protein {Arabidopsis thaliana},
partial (18%)
Length = 463
Score = 52.0 bits (123), Expect(2) = 2e-16
Identities = 31/61 (50%), Positives = 39/61 (63%), Gaps = 3/61 (4%)
Frame = +3
Query: 14 VPLATLIGRELRNGKTEKP--FVKYGQAGLAKKGEDYFLIKTDCHRVPGDS-STSFSVFA 70
VPL+ L+ RE + K EKP V YG A KKGED+ L+KT+C RV D ST F ++A
Sbjct: 198 VPLSVLLKRESLSEKIEKPDMVVVYGNASENKKGEDFILLKTECQRVVCDGVSTLFCLWA 377
Query: 71 I 71
I
Sbjct: 378 I 380
Score = 51.6 bits (122), Expect(2) = 2e-16
Identities = 21/32 (65%), Positives = 28/32 (86%)
Frame = +1
Query: 66 FSVFAILDGHNGISAAIFAKENIINNVMSAIP 97
+SVF + DGHNG +AAI+AKEN++NNV+ AIP
Sbjct: 361 YSVFGLFDGHNGSAAAIYAKENLLNNVLGAIP 456
>TC77609 weakly similar to GP|9909177|dbj|BAB12036.1 putative protein
phosphatase {Oryza sativa (japonica cultivar-group)},
partial (70%)
Length = 1727
Score = 83.2 bits (204), Expect = 2e-16
Identities = 77/250 (30%), Positives = 119/250 (46%), Gaps = 19/250 (7%)
Frame = +2
Query: 69 FAILDGHNGISAAIFAKENIINNVM--SAIPQGVSREEW-LQALPRALVVAFVKTDMEF- 124
+A+ DGH G AA F K N + + + Q +E+ L+ L + AF++ D+
Sbjct: 491 YAVFDGHGGPDAAAFVKRNAMRLFFEDTDMLQSYDTDEFFLEKLEDSHRRAFIRADLALA 670
Query: 125 --QKKGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERER 182
Q + GTTA ++ G + VA+ GD R +L +G V + + DHR + ER R
Sbjct: 671 DEQSVSNSCGTTALTALVLGRHLLVANAGDCRAVLCRRGTAVEM-SKDHR-PSYLPERRR 844
Query: 183 VTASGGEV--GRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDV------GEYIVPIPHVKQ 234
V GG + G LN G L ++R++GD D+ ++ P V+
Sbjct: 845 VEELGGFIDDGYLN--------------GYLSVTRALGDWDLKFPLGAASPLIAEPDVQL 982
Query: 235 VKLSNAGGRLIIASDGIWDTLSSDMAAKSC-----RGVPAELAAKLVVKEALRSRGLKDD 289
V L+ LIIA DGIWD +SS +A R + +A+ +VKEALR D+
Sbjct: 983 VTLTEEDEFLIIACDGIWDVMSSQVAVSFVRRGLRRHSDPQQSARDLVKEALR-LNTSDN 1159
Query: 290 TTCLVVDIIP 299
T +V+ + P
Sbjct: 1160LTAIVICLSP 1189
>TC86518 homologue to PIR|T09640|T09640 protein phosphatase 2C - alfalfa,
partial (50%)
Length = 831
Score = 82.8 bits (203), Expect = 2e-16
Identities = 58/188 (30%), Positives = 96/188 (50%), Gaps = 7/188 (3%)
Frame = +1
Query: 117 FVKTDMEFQKKGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEEN 176
++ TD EF KK G+ I + V++ GD R ++ ++GGV LT DHR
Sbjct: 40 YLNTDSEFMKKDLHGGSCCVTAFIRNGNLVVSNAGDCRAVI-SRGGVAEALTSDHRPSRE 216
Query: 177 VEERERVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVK 236
+E++R+ GG V G R G L +SR IGD + +++ P K ++
Sbjct: 217 -DEKDRIETLGGYVDLCR-------GVWRI-QGSLAVSRGIGDRHLKQWVTAEPETKVIR 369
Query: 237 LSNAGGRLIIASDGIWDTLSS----DMAAKSCRGVPAE---LAAKLVVKEALRSRGLKDD 289
+ LI+ASDG+WD +S+ D+A + C G + +A K + K ++ SRG DD
Sbjct: 370 IEPEHDLLILASDGLWDKVSNQEAVDIARQFCVGNNNQQPLMACKKLAKLSV-SRGSLDD 546
Query: 290 TTCLVVDI 297
T+ +++ +
Sbjct: 547 TSVMIIKL 570
>TC78064 similar to GP|20259167|gb|AAM14299.1 putative phosphatase 2C
{Arabidopsis thaliana}, partial (56%)
Length = 1271
Score = 81.6 bits (200), Expect = 5e-16
Identities = 67/244 (27%), Positives = 119/244 (48%), Gaps = 5/244 (2%)
Frame = +3
Query: 46 EDYFLIKTDCHRVPGDSSTSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEW 105
ED++ IKT S +F I DGH G AA + K+++ N+M ++
Sbjct: 615 EDFYDIKTSTI-----DGRSVCLFGIFDGHGGSRAAEYLKDHLFENLMK-------HPKF 758
Query: 106 LQALPRALVVAFVKTDMEFQKKGETS----GTTATFVIIDGWTVTVASVGDSRCILDTQG 161
L A+ + +TD EF + + G+TA+ ++ + VA+VGDSR ++ G
Sbjct: 759 LTDTKLAISETYQQTDAEFLNSEKDNFRDDGSTASTAVLVDNRLYVANVGDSRTVISKAG 938
Query: 162 GVVSLLTVDHRLEENVEERERVTASGGEVGRLNVFGGN-EVGPLRCWPGGLCLSRSIGDT 220
++L + DH+ + +ER+R+ +GG V ++ G VG G L +SR+ G+
Sbjct: 939 KAIAL-SEDHKPNRS-DERKRIENAGGVV----MWAGTWRVG------GVLAMSRAFGNR 1082
Query: 221 DVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSDMAAKSCRGVPAELAAKLVVKEA 280
+ ++V P ++ ++ L++ASDG+WD + ++ A PAE A+ +
Sbjct: 1083MLKPFVVAEPEIQDQEIDEETEVLVLASDGLWDVVQNEDAVSL---APAEEGAEAAAPKX 1253
Query: 281 LRSR 284
SR
Sbjct: 1254NGSR 1265
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,195,660
Number of Sequences: 36976
Number of extensions: 162253
Number of successful extensions: 763
Number of sequences better than 10.0: 87
Number of HSP's better than 10.0 without gapping: 736
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 741
length of query: 423
length of database: 9,014,727
effective HSP length: 99
effective length of query: 324
effective length of database: 5,354,103
effective search space: 1734729372
effective search space used: 1734729372
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147963.9


