

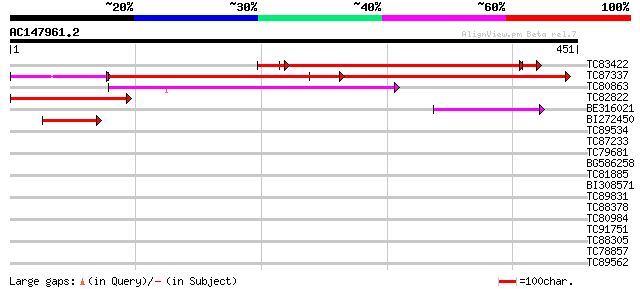
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147961.2 - phase: 0 /pseudo
(451 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC83422 weakly similar to GP|1209703|gb|AAB87721.1| maize gl1 ho... 228 2e-68
TC87337 similar to GP|10716610|gb|AAG21908.1 putative CER1 {Oryz... 240 7e-64
TC80863 similar to GP|498038|gb|AAA33934.1|| lipid transfer prot... 139 2e-33
TC82822 similar to GP|2317909|gb|AAC24373.1| CER1-like protein {... 135 3e-32
BE316021 similar to GP|22900949|gb cuticle protein {Arabidopsis ... 59 4e-09
BI272450 similar to GP|1199467|dbj possible aldehyde decarbonyla... 49 5e-06
TC89534 similar to SP|Q9C5Z1|IF39_ARATH Eukaryotic translation i... 34 0.10
TC87233 similar to PIR|T06048|T06048 kinesin-related protein kat... 31 0.86
TC79681 similar to GP|20453116|gb|AAM19800.1 AT5g22640/MDJ22_6 {... 30 1.9
BG586258 29 3.3
TC81885 similar to GP|11072019|gb|AAG28898.1 F12A21.28 {Arabidop... 29 3.3
BI308571 weakly similar to GP|14018044|gb Conserved hypothetical... 29 4.3
TC89831 similar to PIR|H96569|H96569 unknown protein 54928-5675... 29 4.3
TC88378 similar to GP|18086484|gb|AAL57695.1 At1g07280/F22G5_32 ... 29 4.3
TC80984 similar to GP|20466586|gb|AAM20610.1 unknown protein {Ar... 28 5.6
TC91751 similar to GP|2739382|gb|AAC14505.1| unknown protein {Ar... 28 7.3
TC88305 weakly similar to GP|19920188|gb|AAM08620.1 Putative Glu... 28 7.3
TC78857 weakly similar to SP|O81974|C7D8_SOYBN Cytochrome P450 7... 28 7.3
TC89562 similar to PIR|T45858|T45858 hypothetical protein F3A4.1... 28 9.5
>TC83422 weakly similar to GP|1209703|gb|AAB87721.1| maize gl1 homolog
{Arabidopsis thaliana}, partial (13%)
Length = 683
Score = 228 bits (582), Expect(3) = 2e-68
Identities = 113/196 (57%), Positives = 143/196 (72%), Gaps = 1/196 (0%)
Frame = +3
Query: 215 NLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLPQLKIKVVDGSSLAAATVLN 274
NL ++ EAEL+ KV+SLGL N+ LN E YI R PQLK+K+VDGSSL A VLN
Sbjct: 54 NL*KKQYFEAELSKVKVLSLGLSNQGDLLNRYGELYIKRYPQLKMKIVDGSSLVVAIVLN 233
Query: 275 NIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVVLYKDELKELEQRINT-SKGNLALSP 333
+IPK NQV L G+ +KV++ I NALC++ +V +Y+D+ + L+ R+++ S+ NL
Sbjct: 234 SIPKEENQVFLCGRLDKVSYAIVNALCERGTKVTTMYRDDHENLQLRLSSKSQKNLVFPG 413
Query: 334 FNTPKIWLVGDEWDEYEQMEAPKGSLFIPFSHFPPKKMRKDCFYHYTPAMITPTTFMNSH 393
N+ KIWLVGD+ +E EQ +APKGSLF+PFS FPPKK RKDCFY TPAMITP N H
Sbjct: 414 SNSAKIWLVGDQCEEVEQKKAPKGSLFVPFSQFPPKKFRKDCFYLXTPAMITPPNLANVH 593
Query: 394 SCENWLPRRVMSAWRI 409
SCENWLPRRVMSAWR+
Sbjct: 594 SCENWLPRRVMSAWRL 641
Score = 36.6 bits (83), Expect(3) = 2e-68
Identities = 16/25 (64%), Positives = 18/25 (72%)
Frame = +2
Query: 198 PRFKRQYFSKWQSKTFNNLIEEAIV 222
PRF QY KWQ +T N LIEEAI+
Sbjct: 2 PRFHVQYLFKWQRETLNKLIEEAIL 76
Score = 33.9 bits (76), Expect(3) = 2e-68
Identities = 13/15 (86%), Positives = 15/15 (99%)
Frame = +2
Query: 409 IAGIIHALEGWNVHE 423
IAGI+HALEGW+VHE
Sbjct: 638 IAGILHALEGWDVHE 682
>TC87337 similar to GP|10716610|gb|AAG21908.1 putative CER1 {Oryza sativa},
partial (70%)
Length = 2272
Score = 240 bits (613), Expect = 7e-64
Identities = 112/189 (59%), Positives = 145/189 (76%), Gaps = 1/189 (0%)
Frame = +3
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMRPKESPDVVHLTHLTTFNSI 138
FHSLHHT+FR NYSLFMP+YDYIYGT+DK++D ++E++L R +E+P+VVHLTHLTT SI
Sbjct: 771 FHSLHHTQFRTNYSLFMPLYDYIYGTMDKASDELHESTLKRKEETPNVVHLTHLTTPESI 950
Query: 139 YQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLESNSFKN-LKLQCWLI 197
Y LR GFA+LAS P TSKWYL LMWPFT +SM +TW+ GR F++E N F + L LQ W I
Sbjct: 951 YHLRFGFAALASKPYTSKWYLWLMWPFTAWSMFLTWVYGRTFIVERNRFDDKLNLQTWAI 1130
Query: 198 PRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLPQL 257
P++ QY +WQ + N +IEEAI++A+ G KV+SLGL N+ +LN Y+ R P+L
Sbjct: 1131PKYSVQYSRQWQKVSINKMIEEAILDADKKGIKVVSLGLMNQEEELNIYGGLYVSRHPKL 1310
Query: 258 KIKVVDGSS 266
+KVVDGSS
Sbjct: 1311NVKVVDGSS 1337
Score = 230 bits (586), Expect = 1e-60
Identities = 106/209 (50%), Positives = 148/209 (70%), Gaps = 1/209 (0%)
Frame = +1
Query: 239 KNHQLNERHEHYIGRLPQLKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIAN 298
K L + + + L +++ +LA A V+N+IPKGT QVLLRGK KVA+ +A
Sbjct: 1255 KRKSLTSMEAYMLVGIQSLMLRL*MVVALAVAAVINSIPKGTTQVLLRGKLTKVAYALAY 1434
Query: 299 ALCKKNVQVVVLYKDELKELEQRI-NTSKGNLALSPFNTPKIWLVGDEWDEYEQMEAPKG 357
LC++ VQ+ + D+ +L++ + ++S N+ + T IWLVGD E EQ++APKG
Sbjct: 1435 TLCQQGVQIATMNDDDYVKLKKALMHSSHSNIVNAKSFTQMIWLVGDGLTEEEQLKAPKG 1614
Query: 358 SLFIPFSHFPPKKMRKDCFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALE 417
+LFIP+S FPPKK RKDC YHYTPAM+TPT+ N HSCE+WLPRRVMSAWR+AGI+H LE
Sbjct: 1615 TLFIPYSQFPPKKHRKDCLYHYTPAMLTPTSIENVHSCEDWLPRRVMSAWRVAGIVHCLE 1794
Query: 418 GWNVHECGDTILSTEKVWEASIRHGFQPL 446
WN HECG +++ +KVW ++++HGF+PL
Sbjct: 1795 EWNEHECGYNMINMDKVWPSALKHGFKPL 1881
Score = 60.5 bits (145), Expect = 1e-09
Identities = 37/82 (45%), Positives = 49/82 (59%), Gaps = 1/82 (1%)
Frame = +2
Query: 1 G*PNFVHCTVILHRIHDISYGF*LTMVENRWCDSYSNIA-CWTSRVFILLAS*STSSSLS 59
G*PN V IL + I +++EN W D Y NIA CW+S + +LL + STSSS
Sbjct: 317 G*PNIVQWVTILFGMLYIGRSITSSIMEN*WSD-YCNIASCWSS*ISLLLVTQSTSSSFP 493
Query: 60 ILSLSFSPSFFNCH*ACHLFHS 81
+ +SFS SFF+C+ HL HS
Sbjct: 494 LF*ISFSSSFFHCYGTNHLCHS 559
>TC80863 similar to GP|498038|gb|AAA33934.1|| lipid transfer protein
{Senecio odorus}, partial (75%)
Length = 1474
Score = 139 bits (351), Expect = 2e-33
Identities = 77/235 (32%), Positives = 127/235 (53%), Gaps = 3/235 (1%)
Frame = +1
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMRPKES---PDVVHLTHLTTF 135
+H+LHHT+ N+ LFMP++D + T++K++ ++ET P V L H+
Sbjct: 739 YHNLHHTEKDTNFCLFMPLFDALGNTLNKNSWTLHETLSSGSGNGATVPRFVFLAHMVDI 918
Query: 136 NSIYQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLESNSFKNLKLQCW 195
+S + S +S T++ + P T ++ W+ + F+L + Q W
Sbjct: 919 SSCMHVPFVLRSASSFAYTTRLFFIPCLPVTFLVVMAMWLWSKTFLLSFYYLRGRLHQTW 1098
Query: 196 LIPRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLP 255
++PR QYF + + N IE+AI+ A+ G KVISL NKN LN + ++ + P
Sbjct: 1099VVPRCGFQYFLPFATDGINKQIEQAILRADKMGVKVISLAALNKNESLNGGGKLFVDKHP 1278
Query: 256 QLKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVVL 310
LK++VV G++L AA +LN IP+ +V L G +K+ IA LC+K V+V++L
Sbjct: 1279NLKVRVVHGNTLTAAVILNEIPQDVEEVFLTGATSKLGRAIALYLCQKKVRVLML 1443
>TC82822 similar to GP|2317909|gb|AAC24373.1| CER1-like protein {Arabidopsis
thaliana}, partial (19%)
Length = 580
Score = 135 bits (340), Expect = 3e-32
Identities = 73/98 (74%), Positives = 79/98 (80%), Gaps = 1/98 (1%)
Frame = +3
Query: 1 G*PNFVHCTVILHRIHDISYGF*LTMVENRWCDSYSNIACWTSRVFILLAS*STSSSLSI 60
G* N VHCT IL+ I ISYGF*LTMVENRWCDSYSNIACWTSRVFILLAS*STSSSLSI
Sbjct: 282 G*SNVVHCTRILYSILYISYGF*LTMVENRWCDSYSNIACWTSRVFILLAS*STSSSLSI 461
Query: 61 LSLSFSPSFFNCH*ACHLFHSLHHTKFRRNYSLF-MPM 97
LS+SFSPSFFNCH*A H + F ++LF +PM
Sbjct: 462 LSVSFSPSFFNCH*AYHFCRTPICGAFGHYFTLFAIPM 575
>BE316021 similar to GP|22900949|gb cuticle protein {Arabidopsis thaliana},
partial (20%)
Length = 388
Score = 58.9 bits (141), Expect = 4e-09
Identities = 33/88 (37%), Positives = 41/88 (46%)
Frame = +1
Query: 338 KIWLVGDEWDEYEQMEAPKGSLFIPFSHFPPKKMRKDCFYHYTPAMITPTTFMNSHSCEN 397
K W+VG EQ AP G+ F F P R+DC Y AM P CE
Sbjct: 112 KAWIVGKWITPREQSWAPSGTHFHQFVVPPILSFRRDCTYGDLAAMRLPEDVEGLGCCEY 291
Query: 398 WLPRRVMSAWRIAGIIHALEGWNVHECG 425
+ R V+ A G++H+LEGW HE G
Sbjct: 292 TMDRGVVHACHAGGVVHSLEGWTHHEVG 375
>BI272450 similar to GP|1199467|dbj possible aldehyde decarbonylase
{Arabidopsis thaliana}, partial (18%)
Length = 320
Score = 48.5 bits (114), Expect = 5e-06
Identities = 22/47 (46%), Positives = 32/47 (67%)
Frame = +3
Query: 27 VENRWCDSYSNIACWTSRVFILLAS*STSSSLSILSLSFSPSFFNCH 73
+EN+WC ++ WTS V +LL + STSSS + +SFS SFF+C+
Sbjct: 15 MENKWCGYDNSFT*WTSGVSLLLVTQSTSSSFPLF*ISFSSSFFHCY 155
>TC89534 similar to SP|Q9C5Z1|IF39_ARATH Eukaryotic translation initiation
factor 3 subunit 9 (eIF-3 eta) (eIF3 p110) (eIF3b)
(p82)., partial (31%)
Length = 981
Score = 34.3 bits (77), Expect = 0.10
Identities = 23/49 (46%), Positives = 30/49 (60%)
Frame = -1
Query: 36 SNIACWTSRVFILLAS*STSSSLSILSLSFSPSFFNCH*ACHLFHSLHH 84
S + TS + LAS S+SSSL+ SL FS S F+ +C+ FHSL H
Sbjct: 669 SLVTSTTSSTSMSLASYSSSSSLASPSLRFSLSSFSS--SCNSFHSLAH 529
>TC87233 similar to PIR|T06048|T06048 kinesin-related protein katB -
Arabidopsis thaliana, partial (18%)
Length = 1488
Score = 31.2 bits (69), Expect = 0.86
Identities = 15/42 (35%), Positives = 27/42 (63%)
Frame = -3
Query: 29 NRWCDSYSNIACWTSRVFILLAS*STSSSLSILSLSFSPSFF 70
NR+C+S+++ C ++ V + +A S +SSL +SFS + F
Sbjct: 1384 NRFCNSFTSFFCPSNSVLVSIADISQTSSLFSARVSFSCNSF 1259
>TC79681 similar to GP|20453116|gb|AAM19800.1 AT5g22640/MDJ22_6 {Arabidopsis
thaliana}, partial (42%)
Length = 1682
Score = 30.0 bits (66), Expect = 1.9
Identities = 19/49 (38%), Positives = 25/49 (50%), Gaps = 1/49 (2%)
Frame = -2
Query: 396 ENWLPRRVMSAWRIAGIIHALEGWNVHECGDTILST-EKVWEASIRHGF 443
+ WLP + S + I II GW+V + D ILS E W +R GF
Sbjct: 1612 QRWLPEQTYSIFLILDIIDDKSGWSVKD--DRILSIYEF*WVFRVRLGF 1472
>BG586258
Length = 836
Score = 29.3 bits (64), Expect = 3.3
Identities = 11/23 (47%), Positives = 14/23 (60%)
Frame = -3
Query: 364 SHFPPKKMRKDCFYHYTPAMITP 386
S FP +++CFYH AMI P
Sbjct: 831 SRFPTTDFQRECFYHIPHAMIIP 763
>TC81885 similar to GP|11072019|gb|AAG28898.1 F12A21.28 {Arabidopsis
thaliana}, partial (28%)
Length = 852
Score = 29.3 bits (64), Expect = 3.3
Identities = 22/72 (30%), Positives = 34/72 (46%), Gaps = 10/72 (13%)
Frame = -2
Query: 40 CWTSRVFILLAS*STSSSLSILSLSFSPSFFN-----CH*-----ACHLFHSLHHTKFRR 89
CW S ++ ++ +TSSS S S S S S F+ C * A +FH H F
Sbjct: 224 CWFSHIYSMMIR-NTSSSSSSSSSSTSSSIFSFVSILCC*SIVTIASLVFHKFHSQSFAY 48
Query: 90 NYSLFMPMYDYI 101
N+ L + ++ +
Sbjct: 47 NFLLILENHEIL 12
>BI308571 weakly similar to GP|14018044|gb Conserved hypothetical protein
{Oryza sativa}, partial (4%)
Length = 837
Score = 28.9 bits (63), Expect = 4.3
Identities = 31/124 (25%), Positives = 50/124 (40%), Gaps = 2/124 (1%)
Frame = +1
Query: 129 LTHLTTFNSIYQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLESNSFK 188
L ++ + S Y +G S P + L + W + SMLM + R ++E K
Sbjct: 427 LLQISRYRSCYCRMIGHLKPFSWPLSQVTCLRIFWMIYLPSMLMEHLYARCVIIEDALLK 606
Query: 189 -NLKLQCWLIPRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERH 247
L L W +Y S+ K + + +ISLGL +LN+R+
Sbjct: 607 RELALHQWKFLLL*IKYASRCHWK----------ILLRTSHLLLISLGLMVI*WKLNQRY 756
Query: 248 -EHY 250
+HY
Sbjct: 757 *KHY 768
>TC89831 similar to PIR|H96569|H96569 unknown protein 54928-56750
[imported] - Arabidopsis thaliana, partial (39%)
Length = 897
Score = 28.9 bits (63), Expect = 4.3
Identities = 18/60 (30%), Positives = 34/60 (56%)
Frame = -3
Query: 124 PDVVHLTHLTTFNSIYQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLE 183
P+ ++ +L T +++ L++ SLAS+ + +LHL +P S L+ + R FVL+
Sbjct: 472 PNPMNSENLPTITNLHSLQIH--SLASSSSSLIHHLHLPFPHGSLSNLLVGL*YRCFVLQ 299
>TC88378 similar to GP|18086484|gb|AAL57695.1 At1g07280/F22G5_32
{Arabidopsis thaliana}, partial (38%)
Length = 1687
Score = 28.9 bits (63), Expect = 4.3
Identities = 12/31 (38%), Positives = 16/31 (50%)
Frame = +2
Query: 69 FFNCH*ACHLFHSLHHTKFRRNYSLFMPMYD 99
F NCH +L H LHH + + L P Y+
Sbjct: 248 FCNCHIMFYLIHHLHHKH*LQQFHLHHPNYE 340
>TC80984 similar to GP|20466586|gb|AAM20610.1 unknown protein {Arabidopsis
thaliana}, partial (24%)
Length = 697
Score = 28.5 bits (62), Expect = 5.6
Identities = 15/46 (32%), Positives = 21/46 (45%)
Frame = -1
Query: 353 EAPKGSLFIPFSHFPPKKMRKDCFYHYTPAMITPTTFMNSHSCENW 398
+ P PFS+ K+MRK F T + TT ++S C W
Sbjct: 409 QRPNSLCLTPFSNLSAKRMRKIIFQLGTNIISKLTTRIDSK*CRQW 272
>TC91751 similar to GP|2739382|gb|AAC14505.1| unknown protein {Arabidopsis
thaliana}, partial (38%)
Length = 1200
Score = 28.1 bits (61), Expect = 7.3
Identities = 20/54 (37%), Positives = 29/54 (53%), Gaps = 2/54 (3%)
Frame = -1
Query: 31 WCDSYSNIACWTSRVFILLAS*STSSSLSILSLSFSPSFFNCH--*ACHLFHSL 82
+ S S +AC +S + S ++S SL +LS SFS FF C AC L+ +
Sbjct: 474 FASSESCLACCSSVCALSRLSFNSSISLFVLSSSFST*FFVCSAVSACFLYSGI 313
>TC88305 weakly similar to GP|19920188|gb|AAM08620.1 Putative Glucan 1
3-beta-glucosidase precursor {Oryza sativa (japonica
cultivar-group)}, partial (15%)
Length = 1003
Score = 28.1 bits (61), Expect = 7.3
Identities = 12/38 (31%), Positives = 18/38 (46%)
Frame = +3
Query: 124 PDVVHLTHLTTFNSIYQLRLGFASLASNPQTSKWYLHL 161
P V H+T + T YQL G+ + A W L++
Sbjct: 546 PSVFHMTIVNTLQGEYQLTNGYGNRAPQVMREHWNLYI 659
>TC78857 weakly similar to SP|O81974|C7D8_SOYBN Cytochrome P450 71D8 (EC
1.14.-.-) (P450 CP7). [Soybean] {Glycine max}, partial
(38%)
Length = 1724
Score = 28.1 bits (61), Expect = 7.3
Identities = 15/37 (40%), Positives = 18/37 (48%), Gaps = 8/37 (21%)
Frame = -1
Query: 363 FSHFPPKKMRKDCFYH--------YTPAMITPTTFMN 391
F HFP K+++K C YH Y A TP T N
Sbjct: 1487 FRHFPIKRVKKVCGYHFYCGDS*WYARAYSTPCTEWN 1377
>TC89562 similar to PIR|T45858|T45858 hypothetical protein F3A4.130 -
Arabidopsis thaliana, partial (24%)
Length = 1472
Score = 27.7 bits (60), Expect = 9.5
Identities = 17/68 (25%), Positives = 30/68 (44%)
Frame = -3
Query: 128 HLTHLTTFNSIYQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLESNSF 187
H+ H F+ + + NP TSK Y+ ++W T+ + R L +++F
Sbjct: 249 HVNHWKAFSKLRDI---------NPSTSKTYIFIIWIRTIN------LFQRVAFLHNSTF 115
Query: 188 KNLKLQCW 195
K+ K W
Sbjct: 114 KSKKCSFW 91
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.328 0.139 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,503,615
Number of Sequences: 36976
Number of extensions: 297684
Number of successful extensions: 2274
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 2227
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2266
length of query: 451
length of database: 9,014,727
effective HSP length: 99
effective length of query: 352
effective length of database: 5,354,103
effective search space: 1884644256
effective search space used: 1884644256
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147961.2


