

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147960.7 + phase: 0
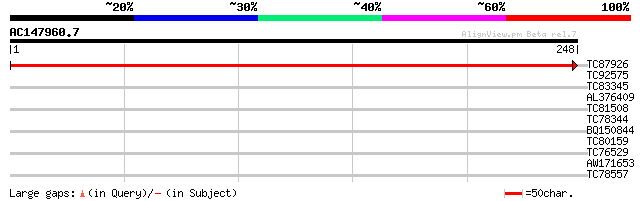
(248 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87926 weakly similar to PIR|F86184|F86184 hypothetical protein... 486 e-138
TC92575 weakly similar to GP|21428434|gb|AAM49877.1 LD13350p {Dr... 32 0.17
TC83345 similar to GP|18086395|gb|AAL57656.1 AT5g13210/T31B5_30 ... 30 1.1
AL376409 weakly similar to GP|19034063|gb hypothetical protein {... 29 1.9
TC81508 similar to PIR|H96662|H96662 hypothetical protein T12P18... 29 1.9
TC78344 similar to GP|13540405|gb|AAK29456.1 histone H1 {Lens cu... 28 4.3
BQ150844 28 4.3
TC80159 similar to PIR|H96730|H96730 unknown protein F5A18.16 [i... 27 7.3
TC76529 similar to PIR|S35757|S35757 vicilin 47K - garden pea, ... 27 9.5
AW171653 27 9.5
TC78557 similar to PIR|A71404|A71404 hypothetical protein - Arab... 27 9.5
>TC87926 weakly similar to PIR|F86184|F86184 hypothetical protein [imported]
- Arabidopsis thaliana, partial (71%)
Length = 931
Score = 486 bits (1250), Expect = e-138
Identities = 248/248 (100%), Positives = 248/248 (100%)
Frame = +2
Query: 1 MSFLAGRLAGKEAAYFFQESKQAVTKLAQKNNPISKTNVVDQRHVVQDNADVLPEVLRHS 60
MSFLAGRLAGKEAAYFFQESKQAVTKLAQKNNPISKTNVVDQRHVVQDNADVLPEVLRHS
Sbjct: 26 MSFLAGRLAGKEAAYFFQESKQAVTKLAQKNNPISKTNVVDQRHVVQDNADVLPEVLRHS 205
Query: 61 LPSKLFRDETASSSSFSASKWVLQSDPKLRSSVSPDAINPLRAFVSLPQVTFGARRWELP 120
LPSKLFRDETASSSSFSASKWVLQSDPKLRSSVSPDAINPLRAFVSLPQVTFGARRWELP
Sbjct: 206 LPSKLFRDETASSSSFSASKWVLQSDPKLRSSVSPDAINPLRAFVSLPQVTFGARRWELP 385
Query: 121 EAKHGVSASTANELRQDRYDVNVNPEKLKAASEGLANLGKAFAIATAVVFGGAAMVIGMV 180
EAKHGVSASTANELRQDRYDVNVNPEKLKAASEGLANLGKAFAIATAVVFGGAAMVIGMV
Sbjct: 386 EAKHGVSASTANELRQDRYDVNVNPEKLKAASEGLANLGKAFAIATAVVFGGAAMVIGMV 565
Query: 181 ASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLEREDVKQKAIVKD 240
ASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLEREDVKQKAIVKD
Sbjct: 566 ASKLELHNMGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLEREDVKQKAIVKD 745
Query: 241 LSKILGSK 248
LSKILGSK
Sbjct: 746 LSKILGSK 769
>TC92575 weakly similar to GP|21428434|gb|AAM49877.1 LD13350p {Drosophila
melanogaster}, partial (3%)
Length = 557
Score = 32.3 bits (72), Expect = 0.17
Identities = 20/50 (40%), Positives = 28/50 (56%)
Frame = +1
Query: 57 LRHSLPSKLFRDETASSSSFSASKWVLQSDPKLRSSVSPDAINPLRAFVS 106
L SLPS++ AS+S F AS L S SS++ + NPL +F+S
Sbjct: 334 LPSSLPSRILSSSMASNSGFLASTLSLSSLLSSVSSLNLSSSNPLSSFLS 483
>TC83345 similar to GP|18086395|gb|AAL57656.1 AT5g13210/T31B5_30
{Arabidopsis thaliana}, partial (45%)
Length = 1085
Score = 29.6 bits (65), Expect = 1.1
Identities = 12/32 (37%), Positives = 19/32 (58%)
Frame = +1
Query: 88 KLRSSVSPDAINPLRAFVSLPQVTFGARRWEL 119
++R + D + PLR + LP+V GA +W L
Sbjct: 331 RVRDRLRKDVLVPLRKVLELPEVFIGANQWGL 426
>AL376409 weakly similar to GP|19034063|gb hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (1%)
Length = 478
Score = 28.9 bits (63), Expect = 1.9
Identities = 22/74 (29%), Positives = 37/74 (49%), Gaps = 5/74 (6%)
Frame = -1
Query: 35 SKTNVVDQRHVVQDN-----ADVLPEVLRHSLPSKLFRDETASSSSFSASKWVLQSDPKL 89
SKT+ + R + DN LPE SKL +D++A+++ S ++ S+PK+
Sbjct: 466 SKTSRLSIRSPLNDNKLSQSVSCLPE-------SKLEKDDSATNTKVSMARIRRLSEPKM 308
Query: 90 RSSVSPDAINPLRA 103
S ++ PL A
Sbjct: 307 SSVRQTSSVKPLGA 266
>TC81508 similar to PIR|H96662|H96662 hypothetical protein T12P18.20
[imported] - Arabidopsis thaliana, partial (65%)
Length = 717
Score = 28.9 bits (63), Expect = 1.9
Identities = 28/105 (26%), Positives = 49/105 (46%)
Frame = +2
Query: 8 LAGKEAAYFFQESKQAVTKLAQKNNPISKTNVVDQRHVVQDNADVLPEVLRHSLPSKLFR 67
L GKE ++ E+K+ + + ++ PI T + ++ ++ D+ E ++P
Sbjct: 185 LEGKERIHY--ENKRKIREALREGKPIP-TELRNEEAALRREIDLEDE--NTAVPRSHID 349
Query: 68 DETASSSSFSASKWVLQSDPKLRSSVSPDAINPLRAFVSLPQVTF 112
DE A F+A K DPK+ + S D PL+ FV + F
Sbjct: 350 DEYA----FAAEK-----DPKILLTTSRDPSAPLKQFVKELSIVF 457
>TC78344 similar to GP|13540405|gb|AAK29456.1 histone H1 {Lens culinaris},
partial (86%)
Length = 1215
Score = 27.7 bits (60), Expect = 4.3
Identities = 14/35 (40%), Positives = 19/35 (54%)
Frame = -1
Query: 150 AASEGLANLGKAFAIATAVVFGGAAMVIGMVASKL 184
A + GLA G A A+A A+ GAA + + A L
Sbjct: 648 ALAAGLALAGAALALALALALAGAAFALDLAAGAL 544
>BQ150844
Length = 546
Score = 27.7 bits (60), Expect = 4.3
Identities = 10/21 (47%), Positives = 15/21 (70%)
Frame = -1
Query: 31 NNPISKTNVVDQRHVVQDNAD 51
NNP+S + ++D+R QDN D
Sbjct: 426 NNPLSISRLLDRRRTTQDNTD 364
>TC80159 similar to PIR|H96730|H96730 unknown protein F5A18.16 [imported]
- Arabidopsis thaliana, partial (98%)
Length = 887
Score = 26.9 bits (58), Expect = 7.3
Identities = 16/35 (45%), Positives = 19/35 (53%)
Frame = +1
Query: 60 SLPSKLFRDETASSSSFSASKWVLQSDPKLRSSVS 94
SLP L SSS S+SKWVL+ P L +S
Sbjct: 37 SLPLFLSLQFQNPSSSDSSSKWVLKDPPSLFLGIS 141
>TC76529 similar to PIR|S35757|S35757 vicilin 47K - garden pea, partial
(94%)
Length = 1588
Score = 26.6 bits (57), Expect = 9.5
Identities = 24/83 (28%), Positives = 38/83 (44%), Gaps = 4/83 (4%)
Frame = +1
Query: 2 SFLAGRLAGKEAAYFFQESKQAVTKLAQKNNPISKTNVVDQRHVVQDNADVLPEVLRHSL 61
SF +G A F ++ L ++ P + + D+RH ADV+ +V R +
Sbjct: 538 SFFSGFSKNILEAAFNANYEEIERVLIEEQEPQHRRGLRDRRHKQSQEADVIVKVSREQI 717
Query: 62 PSKLFRDETASS----SSFSASK 80
+L R +SS SS SAS+
Sbjct: 718 -EELSRHAKSSSRRSASSESASR 783
>AW171653
Length = 390
Score = 26.6 bits (57), Expect = 9.5
Identities = 13/22 (59%), Positives = 15/22 (68%)
Frame = +1
Query: 54 PEVLRHSLPSKLFRDETASSSS 75
PEVL +SL S LF ASSS+
Sbjct: 73 PEVLNYSLHSNLFTPSAASSSA 138
>TC78557 similar to PIR|A71404|A71404 hypothetical protein - Arabidopsis
thaliana, partial (72%)
Length = 1690
Score = 26.6 bits (57), Expect = 9.5
Identities = 14/57 (24%), Positives = 27/57 (46%)
Frame = +1
Query: 187 HNMGDLKTKGKDVVEPQLENIKNYFVPMKVWAENMSRKWHLEREDVKQKAIVKDLSK 243
H +K KGK PQ+ + + + E+ L+++DVK + +V D+ +
Sbjct: 1042 HMAAVVKAKGKGKETPQIIDEEKFDAKKSAGGESQLTTPLLQKQDVKSENVVVDIDE 1212
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,345,742
Number of Sequences: 36976
Number of extensions: 76101
Number of successful extensions: 367
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 367
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 367
length of query: 248
length of database: 9,014,727
effective HSP length: 94
effective length of query: 154
effective length of database: 5,538,983
effective search space: 853003382
effective search space used: 853003382
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 57 (26.6 bits)
Medicago: description of AC147960.7


