

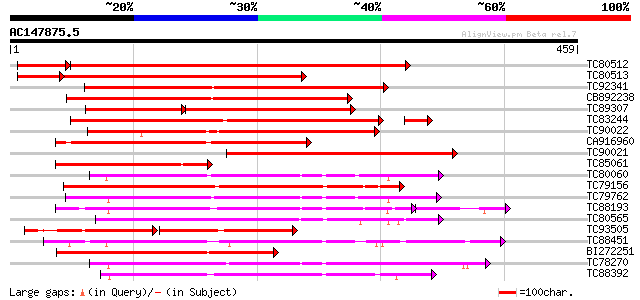
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147875.5 - phase: 0
(459 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC80512 weakly similar to PIR|A96693|A96693 probable receptor se... 547 e-177
TC80513 weakly similar to GP|9755449|gb|AAF98210.1| Unknown prot... 367 e-117
TC92341 similar to GP|21741394|emb|CAD39676. OSJNBb0051N19.7 {Or... 336 1e-92
CB892238 weakly similar to GP|21741394|em OSJNBb0051N19.7 {Oryza... 318 4e-87
TC89307 similar to GP|21741394|emb|CAD39676. OSJNBb0051N19.7 {Or... 194 1e-78
TC83244 weakly similar to GP|11034602|dbj|BAB17126. putative rec... 272 5e-78
TC90022 similar to GP|14587271|dbj|BAB61189. putative receptor-l... 268 4e-72
CA916960 weakly similar to GP|14090203|db putative receptor kina... 223 1e-58
TC90021 weakly similar to GP|13377502|gb|AAK20740.1 LRK33 {Triti... 215 3e-56
TC85061 similar to GP|9755449|gb|AAF98210.1| Unknown protein {Ar... 205 3e-53
TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.... 204 8e-53
TC79156 weakly similar to GP|11034603|dbj|BAB17127. putative rec... 202 2e-52
TC79762 similar to PIR|H96731|H96731 hypothetical protein F5A18.... 196 1e-50
TC88193 similar to PIR|G96602|G96602 probable receptor protein k... 183 2e-50
TC80565 weakly similar to GP|14335072|gb|AAK59800.1 AT5g20050/F2... 196 2e-50
TC93505 weakly similar to GP|15408735|dbj|BAB64138. putative rec... 134 4e-48
TC88451 similar to GP|836954|gb|AAC23542.1|| receptor protein ki... 184 5e-47
BI272251 similar to GP|13377503|gb TAK33 {Triticum aestivum}, pa... 184 6e-47
TC78270 similar to GP|16649103|gb|AAL24403.1 Unknown protein {Ar... 181 4e-46
TC88392 similar to GP|11275527|dbj|BAB18292. putative receptor-l... 181 5e-46
>TC80512 weakly similar to PIR|A96693|A96693 probable receptor
serine/threonine kinase PR5K T4O24.7 [imported] -
Arabidopsis thaliana, partial (35%)
Length = 1077
Score = 547 bits (1410), Expect(2) = e-177
Identities = 271/275 (98%), Positives = 272/275 (98%)
Frame = +1
Query: 50 VEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESK 109
VEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESK
Sbjct: 253 VEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESK 432
Query: 110 GNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVC 169
GNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSL+KFIYKSGFPDAVC
Sbjct: 433 GNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLNKFIYKSGFPDAVC 612
Query: 170 DCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQM 229
D DSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQM
Sbjct: 613 DFDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQM 792
Query: 230 NDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTS 289
NDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTS
Sbjct: 793 NDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTS 972
Query: 290 EMYFPDWIYKDLEQGNDLLNSLTISEEENDMVKKI 324
EMYFPDWIYKDLEQGNDL NSLTISEEENDM KKI
Sbjct: 973 EMYFPDWIYKDLEQGNDLXNSLTISEEENDMXKKI 1077
Score = 92.0 bits (227), Expect(2) = e-177
Identities = 43/43 (100%), Positives = 43/43 (100%)
Frame = +3
Query: 7 GASVAGFGVTMFFIIFISCYFKKGIRRPQMTIFRKRRKHVDNN 49
GASVAGFGVTMFFIIFISCYFKKGIRRPQMTIFRKRRKHVDNN
Sbjct: 123 GASVAGFGVTMFFIIFISCYFKKGIRRPQMTIFRKRRKHVDNN 251
>TC80513 weakly similar to GP|9755449|gb|AAF98210.1| Unknown protein
{Arabidopsis thaliana}, partial (14%)
Length = 824
Score = 367 bits (942), Expect(2) = e-117
Identities = 183/201 (91%), Positives = 190/201 (94%), Gaps = 1/201 (0%)
Frame = +2
Query: 41 KRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQV 100
K KHVD+NVEVFMQSYNLSIARRYSY EVKRITNSFRDKLGHGGYGVVYKASLTDGRQV
Sbjct: 218 KEEKHVDSNVEVFMQSYNLSIARRYSYTEVKRITNSFRDKLGHGGYGVVYKASLTDGRQV 397
Query: 101 AVKVINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIY 160
AVKVINESKGNGEEFINEVASISRTSH+NIVSLLG+CYE NKRALIYE+MPKGSLDKFIY
Sbjct: 398 AVKVINESKGNGEEFINEVASISRTSHLNIVSLLGFCYEVNKRALIYEYMPKGSLDKFIY 577
Query: 161 KSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISD 220
KSGFPDAVCD D NTLFQ+AIGIARGLEYLHQGCSSRILHLDIKPQNILLDE+FCPKISD
Sbjct: 578 KSGFPDAVCDFDWNTLFQVAIGIARGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISD 757
Query: 221 FGLAKICQMNDSI-VSIPGTR 240
FGLAKICQ DSI V+I TR
Sbjct: 758 FGLAKICQRKDSICVNIGDTR 820
Score = 73.9 bits (180), Expect(2) = e-117
Identities = 35/38 (92%), Positives = 36/38 (94%)
Frame = +1
Query: 7 GASVAGFGVTMFFIIFISCYFKKGIRRPQMTIFRKRRK 44
GASVAGFGVTMFFII ISCYFKKGIRR +MTIFRKRRK
Sbjct: 115 GASVAGFGVTMFFIIMISCYFKKGIRRQEMTIFRKRRK 228
>TC92341 similar to GP|21741394|emb|CAD39676. OSJNBb0051N19.7 {Oryza
sativa}, partial (60%)
Length = 779
Score = 336 bits (862), Expect = 1e-92
Identities = 159/246 (64%), Positives = 198/246 (79%)
Frame = +2
Query: 61 IARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVA 120
+ +RY Y+E+K +TNS +DKLG GG+GVVYK L +G VAVK++N SKGNGEEFINEVA
Sbjct: 20 LQKRYKYSEIKNMTNSLKDKLGQGGFGVVYKGKLFNGCLVAVKILNVSKGNGEEFINEVA 199
Query: 121 SISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIA 180
SISRTSH+N+V+LLG+C+E NK+AL+YEFM GSLDKFIY + + + L++IA
Sbjct: 200 SISRTSHVNVVTLLGFCFEGNKKALVYEFMSNGSLDKFIYNKEL-ETIASLSWDKLYKIA 376
Query: 181 IGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTR 240
GIARGLEYLH GC++RILH DIKP NILLD+N CPKISDFGLAK+C +SIVS+ R
Sbjct: 377 KGIARGLEYLHGGCTTRILHFDIKPHNILLDDNLCPKISDFGLAKLCLRKESIVSMSDQR 556
Query: 241 GTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYKD 300
GT+GY+APEV++R FGGVS+KSDVYSYGM++LEM+GGRKN S TSE+YFP W+YK
Sbjct: 557 GTMGYVAPEVWNRHFGGVSHKSDVYSYGMILLEMVGGRKNINADASRTSEIYFPHWVYKR 736
Query: 301 LEQGND 306
LE +D
Sbjct: 737 LELASD 754
>CB892238 weakly similar to GP|21741394|em OSJNBb0051N19.7 {Oryza sativa},
partial (61%)
Length = 726
Score = 318 bits (814), Expect = 4e-87
Identities = 146/231 (63%), Positives = 189/231 (81%)
Frame = +3
Query: 47 DNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVIN 106
D N+E F++ + + +RY Y+E+K++T+SF+ KLG GG+GVVYK L +G VA+K++N
Sbjct: 36 DTNIEAFLKDHGALLQKRYKYSEIKKMTDSFKVKLGQGGFGVVYKGKLFNGCHVAIKILN 215
Query: 107 ESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPD 166
SKGNGEEFINEV+SI+RTSH+N+V+LLG+C+E K+ALIYEFM GSLDKFIY G P+
Sbjct: 216 SSKGNGEEFINEVSSITRTSHVNVVTLLGFCFEGTKKALIYEFMSNGSLDKFIYNKG-PE 392
Query: 167 AVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKI 226
+ L+QIA GIARGLEYLH+GC++RILH DIKP NILLDEN CPKISDFGLAK+
Sbjct: 393 TIASLSWENLYQIAKGIARGLEYLHRGCTTRILHFDIKPHNILLDENLCPKISDFGLAKL 572
Query: 227 CQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGG 277
C +SI+S+ RGT+GY+APEV++R FGGVS+KSDVYSYGM++LEM+GG
Sbjct: 573 CPKQESIISMSDQRGTMGYVAPEVWNRHFGGVSHKSDVYSYGMMLLEMVGG 725
>TC89307 similar to GP|21741394|emb|CAD39676. OSJNBb0051N19.7 {Oryza
sativa}, partial (58%)
Length = 814
Score = 194 bits (494), Expect(2) = 1e-78
Identities = 88/139 (63%), Positives = 117/139 (83%), Gaps = 1/139 (0%)
Frame = +2
Query: 143 RALIYEFMPKGSLDKFIYKSGFPDAV-CDCDSNTLFQIAIGIARGLEYLHQGCSSRILHL 201
+ALIY++MP GSL+KFIY++ P + T++ IAIG+ARGLEYLH+GC+++ILH
Sbjct: 398 KALIYDYMPNGSLEKFIYENKDPLKLNLQLSCKTVYNIAIGVARGLEYLHKGCNTKILHF 577
Query: 202 DIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYK 261
DIKP NILLD++FCPK+SDFGLAK+C +SI+S+ G RGT GY+APEVFSR FGGVS+K
Sbjct: 578 DIKPHNILLDDDFCPKVSDFGLAKVCPRKESIISLLGARGTAGYIAPEVFSRNFGGVSHK 757
Query: 262 SDVYSYGMLILEMIGGRKN 280
SDVY+YGM++LEM+GG++N
Sbjct: 758 SDVYNYGMMVLEMVGGKQN 814
Score = 117 bits (292), Expect(2) = 1e-78
Identities = 57/82 (69%), Positives = 68/82 (82%)
Frame = +3
Query: 62 ARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVAS 121
A+RY+YAE+K+ TNSF+ KLG GGYG VYK L DG VAVKV++ES+GNGEEFINEVAS
Sbjct: 156 AKRYTYAEIKKATNSFKYKLGQGGYGSVYKGKLQDGSLVAVKVLSESEGNGEEFINEVAS 335
Query: 122 ISRTSHMNIVSLLGYCYEANKR 143
IS TSH+NIV LLG+ E +KR
Sbjct: 336 ISVTSHVNIVGLLGFYLEGSKR 401
>TC83244 weakly similar to GP|11034602|dbj|BAB17126. putative receptor
kinase {Oryza sativa (japonica cultivar-group)}, partial
(36%)
Length = 1366
Score = 272 bits (695), Expect(2) = 5e-78
Identities = 127/253 (50%), Positives = 177/253 (69%)
Frame = +1
Query: 50 VEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESK 109
+E F++ Y R++YA++KRITN F++ LG G +G V+K L+ VAVKV+NE++
Sbjct: 496 IEKFLEDYRALKPTRFTYADIKRITNGFKESLGEGAHGSVFKGMLSQEILVAVKVLNETQ 675
Query: 110 GNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVC 169
G+G +FINEV ++ + H+N+V LLG+C + RAL+Y+F P GSL F+ +
Sbjct: 676 GDGNDFINEVGTMGKIHHVNVVRLLGFCADGFHRALVYDFFPNGSLQNFLAPPENKEVFL 855
Query: 170 DCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQM 229
+ L +IA+G+ARG+EYLH GC RILH DI P N+L+D+N PKI+DFGLAK+C
Sbjct: 856 GREK--LQRIALGVARGIEYLHIGCDHRILHFDINPHNVLIDDNLSPKITDFGLAKLCPK 1029
Query: 230 NDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTS 289
N S VS+ RGT+GY+APEVFSR FG VSYKSD+YSYGML+LEM+GGRKN T
Sbjct: 1030NQSTVSMTAARGTLGYIAPEVFSRNFGNVSYKSDIYSYGMLLLEMVGGRKNTNQSAKETF 1209
Query: 290 EMYFPDWIYKDLE 302
++ +P+WI+ +E
Sbjct: 1210QVLYPEWIHNLIE 1248
Score = 37.7 bits (86), Expect(2) = 5e-78
Identities = 14/23 (60%), Positives = 18/23 (77%)
Frame = +2
Query: 320 MVKKITMVSLWCIQTNPLDRPPM 342
+ KK+ +V LWCIQ NP+DRP M
Sbjct: 1295 IAKKLALVGLWCIQWNPVDRPSM 1363
>TC90022 similar to GP|14587271|dbj|BAB61189. putative receptor-like kinase
{Oryza sativa (japonica cultivar-group)}, partial (31%)
Length = 773
Score = 268 bits (684), Expect = 4e-72
Identities = 126/238 (52%), Positives = 176/238 (73%), Gaps = 2/238 (0%)
Frame = +1
Query: 64 RYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVI--NESKGNGEEFINEVAS 121
RYSY E+K++TN F+DKLG GGYG VYK L G VA+K++ ++ KGNG++FINEVA+
Sbjct: 13 RYSYTEIKKMTNGFKDKLGEGGYGKVYKGKLRSGPSVAIKMLGKHKGKGNGQDFINEVAT 192
Query: 122 ISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAI 181
I R H N+V L+G+C E +KRAL+Y+FMP GSLDK+I S D + +++I++
Sbjct: 193 IGRIHHTNVVRLIGFCVEGSKRALVYDFMPNGSLDKYI--SSREDHI-SLTYKQIYEISL 363
Query: 182 GIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRG 241
+ARG+ YLHQGC +ILH DIKP NILLD++F K+SDFGLAK+ + +SIV++ RG
Sbjct: 364 AVARGIAYLHQGCDMQILHFDIKPHNILLDQDFIAKVSDFGLAKLYPIENSIVTMTAARG 543
Query: 242 TIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYK 299
TIGYMAPE+F + G VSYK+DVYS+GML++E+ R+N + +S+++FP WIY+
Sbjct: 544 TIGYMAPELFYKNIGKVSYKADVYSFGMLLMEIANRRRNLNSNADDSSQIFFPYWIYQ 717
>CA916960 weakly similar to GP|14090203|db putative receptor kinase {Oryza
sativa (japonica cultivar-group)}, partial (27%)
Length = 651
Score = 223 bits (568), Expect = 1e-58
Identities = 112/208 (53%), Positives = 145/208 (68%), Gaps = 1/208 (0%)
Frame = +2
Query: 38 IFRKRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDG 97
I RK ++ + +E F++ Y RYSY E+KRITN+F D LG G YG VYK S++
Sbjct: 44 ILRKEKQAI---IEKFLEDYRALKPTRYSYVEIKRITNNFGDMLGQGAYGTVYKGSISKE 214
Query: 98 RQVAVKVINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDK 157
VAVK++N S+GNG++F+NEV ++ R H+NIV L+G+C + KRALIYEF+P GSL K
Sbjct: 215 FSVAVKILNVSQGNGQDFLNEVGTMGRIHHVNIVRLIGFCADGFKRALIYEFLPNGSLQK 394
Query: 158 FIYKSGFPDAVCD-CDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCP 216
FI PD + L +IA+GIA+G+EYLHQGC RILH DIKPQN+LLD NF P
Sbjct: 395 FI---NSPDNKKNFLGWKKLHEIALGIAKGIEYLHQGCDQRILHFDIKPQNVLLDHNFIP 565
Query: 217 KISDFGLAKICQMNDSIVSIPGTRGTIG 244
KI DFGLAK+C + SIVS+ RGT+G
Sbjct: 566 KICDFGLAKLCSRDQSIVSMTAARGTLG 649
>TC90021 weakly similar to GP|13377502|gb|AAK20740.1 LRK33 {Triticum
aestivum}, partial (23%)
Length = 736
Score = 215 bits (547), Expect = 3e-56
Identities = 95/187 (50%), Positives = 141/187 (74%)
Frame = +3
Query: 176 LFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVS 235
+++I++G+ARG+ YLHQGC +ILH DIKP NILLD++F K+SDFGLAK+ +++SIV+
Sbjct: 54 MYEISLGVARGIAYLHQGCDMQILHFDIKPHNILLDQDFIAKVSDFGLAKLYPVDNSIVT 233
Query: 236 IPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPD 295
+ RGTIGYMAPE+F + G VSYK+DVYS+GML++E+ R+N + +S+++FP
Sbjct: 234 LTAARGTIGYMAPELFYKNIGKVSYKADVYSFGMLLMEIANRRRNLNSNADDSSQIFFPY 413
Query: 296 WIYKDLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSS 355
WIY +L + ++ S ++EE VKK+ +++LWCIQ NP+DRP M++VIEML+G +
Sbjct: 414 WIYNELMEEREIDISGEATDEEKKNVKKMFIIALWCIQLNPIDRPSMDRVIEMLEGDIED 593
Query: 356 VTFPPKP 362
+ PPKP
Sbjct: 594 IEIPPKP 614
>TC85061 similar to GP|9755449|gb|AAF98210.1| Unknown protein {Arabidopsis
thaliana}, partial (8%)
Length = 1013
Score = 205 bits (521), Expect = 3e-53
Identities = 103/128 (80%), Positives = 116/128 (90%), Gaps = 1/128 (0%)
Frame = +1
Query: 38 IFRKRRKHVDNNVEVFMQSYNLSIA-RRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTD 96
I RK K +DNNVE F+QSYNLS+ ++Y YAEVK++TNSFRDKLG GGYGVVYKA+L D
Sbjct: 628 ILRKTTKSIDNNVEDFIQSYNLSVPIKQYRYAEVKKMTNSFRDKLGQGGYGVVYKANLPD 807
Query: 97 GRQVAVKVINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLD 156
GRQVAVK+INESKGNGE+FINEVASISRTSH+NIVSLLG+CYE NKRALIYEF+PKGSLD
Sbjct: 808 GRQVAVKIINESKGNGEDFINEVASISRTSHVNIVSLLGFCYE-NKRALIYEFLPKGSLD 984
Query: 157 KFIYKSGF 164
KFI KSGF
Sbjct: 985 KFILKSGF 1008
>TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.24
[imported] - Arabidopsis thaliana, partial (32%)
Length = 1547
Score = 204 bits (518), Expect = 8e-53
Identities = 120/293 (40%), Positives = 178/293 (59%), Gaps = 6/293 (2%)
Frame = +1
Query: 65 YSYAEVKRITNSF--RDKLGHGGYGVVYKASLTDGRQVAVKVIN-ESKGNGEEFINEVAS 121
YS ++K TN+F ++K+G GG+G VYK L+DG +AVK ++ +SK EF+NE+
Sbjct: 157 YSLRQIKVATNNFDPKNKIGEGGFGPVYKGVLSDGAVIAVKQLSSKSKQGNREFVNEIGM 336
Query: 122 ISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAI 181
IS H N+V L G C E N+ L+YE+M SL + ++ G P+ + D T +I +
Sbjct: 337 ISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALF--GKPEQRLNLDWRTRMKICV 510
Query: 182 GIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRG 241
GIARGL YLH+ +I+H DIK N+LLD+N KISDFGLAK+ + ++ +S G
Sbjct: 511 GIARGLAYLHEESRLKIVHRDIKATNVLLDKNLNAKISDFGLAKLDEEENTHIS-TRIAG 687
Query: 242 TIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSE-MYFPDWIYKD 300
TIGYMAPE R G ++ K+DVYS+G++ LE++ G N T E +Y DW Y
Sbjct: 688 TIGYMAPEYAMR--GYLTDKADVYSFGVVALEIVSGMSN--TNYRPKEEFVYLLDWAYVL 855
Query: 301 LEQGN--DLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQG 351
EQGN +L++ S+ ++ ++ ++L C +P RPPM+ V+ ML+G
Sbjct: 856 QEQGNLLELVDPTLGSKYSSEEAMRMLQLALLCTNPSPTLRPPMSSVVSMLEG 1014
>TC79156 weakly similar to GP|11034603|dbj|BAB17127. putative receptor
kinase {Oryza sativa (japonica cultivar-group)}, partial
(18%)
Length = 1246
Score = 202 bits (514), Expect = 2e-52
Identities = 115/278 (41%), Positives = 171/278 (61%), Gaps = 2/278 (0%)
Frame = +1
Query: 44 KHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVK 103
KH +E + N R++ ++ IT + LG G +GVV+K L++G VAVK
Sbjct: 427 KHEFATMERIFSNINREKPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVK 606
Query: 104 VIN-ESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKS 162
V+N G E+F EV +I RT H+N+V L G+C+ +KRAL+YE++ GSLDK+I+ S
Sbjct: 607 VLNCLDMGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGS 786
Query: 163 GFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFG 222
+ D D L +IAIG A+G+ YLH+ C RI+H DIKP+N+LLD PKI+DFG
Sbjct: 787 KNRN---DFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFG 957
Query: 223 LAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQ 282
LAK+ +I RGT GY APE++ V+YK DVYS+G+L+ E++G R+++
Sbjct: 958 LAKLRSRESNIELNTHFRGTRGYAAPEMWKPY--PVTYKCDVYSFGILLFEIVGRRRHFD 1131
Query: 283 TGGSCTSEMYFPDWIYKDLEQGNDLLNSLTISE-EEND 319
+ S S+ +FP W + ++ + N+L+ L + E EE D
Sbjct: 1132SSYS-ESQQWFPRWTW-EMFENNELVVMLALCEIEEKD 1239
>TC79762 similar to PIR|H96731|H96731 hypothetical protein F5A18.8
[imported] - Arabidopsis thaliana, partial (55%)
Length = 1564
Score = 196 bits (499), Expect = 1e-50
Identities = 116/309 (37%), Positives = 178/309 (57%), Gaps = 5/309 (1%)
Frame = +3
Query: 46 VDNNVEVFMQSYNLSIARRYSYAEVKRITNSFR--DKLGHGGYGVVYKASLTDGRQVAVK 103
V+ + E +Q + +SY + T +F KLG GG+G VYK L+DGR+VAVK
Sbjct: 291 VEGDNEAELQKMASREQKIFSYETLLSATKNFNATHKLGEGGFGPVYKGKLSDGREVAVK 470
Query: 104 VINESKGNGE-EFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKS 162
++++ G+ EF+NE ++R H N+V+LLGYC ++ L+YE++P SLDKF++K
Sbjct: 471 KLSQTSNQGKKEFMNEAKLLARVQHKNVVNLLGYCVHGTEKILVYEYVPHESLDKFLFKE 650
Query: 163 GFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFG 222
+ D F I G+A+GL YLH+ + I+H DIK NILLD+ + KI+DFG
Sbjct: 651 A--EKREQLDWKRRFGIITGVAKGLLYLHEDSHNCIIHRDIKASNILLDDKWTAKIADFG 824
Query: 223 LAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQ 282
+A++ + S V GT GYMAPE G +S K+DV+SYG+L+LE+I G++N
Sbjct: 825 MARLFPEDQSQVK-TRVAGTNGYMAPEYMMH--GRLSVKADVFSYGVLVLELITGQRN-S 992
Query: 283 TGGSCTSEMYFPDWIYKDLEQGN--DLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRP 340
+ E DW YK ++G ++++S S + V ++L CIQ +P RP
Sbjct: 993 SFNLXVEEHNLLDWAYKMYKKGRSLEIVDSALASTVLTEQVDMCIQLALLCIQGDPQLRP 1172
Query: 341 PMNKVIEML 349
M +++ L
Sbjct: 1173TMRRIVVKL 1199
>TC88193 similar to PIR|G96602|G96602 probable receptor protein kinase
F14G9.24 [imported] - Arabidopsis thaliana, partial
(20%)
Length = 1815
Score = 183 bits (465), Expect(2) = 2e-50
Identities = 112/297 (37%), Positives = 177/297 (58%), Gaps = 5/297 (1%)
Frame = +3
Query: 38 IFRKRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFR--DKLGHGGYGVVYKASLT 95
I R+R+ + D++ V + + + +SY E+K T+ F +KLG GG+G VYK +L
Sbjct: 471 ILRRRKLYNDDDDLVGIDT----MPNTFSYYELKNATSDFNRDNKLGEGGFGPVYKGTLN 638
Query: 96 DGRQVAVKVINESKGNGE-EFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGS 154
DGR VAVK ++ G+ +FI E+A+IS H N+V L G C E NKR L+YE++ S
Sbjct: 639 DGRFVAVKQLSIGSHQGKSQFIAEIATISAVQHRNLVKLYGCCIEGNKRLLVYEYLENKS 818
Query: 155 LDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENF 214
LD+ ++ + V + +T + + +G+ARGL YLH+ RI+H D+K NILLD
Sbjct: 819 LDQALFGN-----VLFLNWSTRYDVCMGVARGLTYLHEESRLRIVHRDVKASNILLDSEL 983
Query: 215 CPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEM 274
PK+SDFGLAK+ + +S GTIGY+APE R G ++ K+DV+S+G++ LE+
Sbjct: 984 VPKLSDFGLAKLYDDKKTHIS-TRVAGTIGYLAPEYAMR--GRLTEKADVFSFGVVALEL 1154
Query: 275 IGGRKNYQTGGSCTSEMYFPDWIYKDLEQG--NDLLNSLTISEEENDMVKKITMVSL 329
+ GR N + +MY DW ++ E+ NDL++ +SE + V+++ + +
Sbjct: 1155VSGRPNSDSSLE-EDKMYLLDWAWQLHERNCINDLIDP-RLSEFNMEEVERLVGIGI 1319
Score = 34.3 bits (77), Expect(2) = 2e-50
Identities = 23/79 (29%), Positives = 36/79 (45%), Gaps = 2/79 (2%)
Frame = +1
Query: 329 LWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKPVLFSPKRPPLQLSNMSSSDWQ--ETN 386
L C QT+P RP M++V+ ML G + T +P + +DW+ + +
Sbjct: 1318 LLCTQTSPNLRPSMSRVVAMLLGDIEVSTVTSRPEYW--------------TDWKFGDVS 1455
Query: 387 SITTETELEEEPIEGYQSS 405
SI T+T E Y S+
Sbjct: 1456 SIMTDTSAEGLDTSNYNST 1512
>TC80565 weakly similar to GP|14335072|gb|AAK59800.1 AT5g20050/F28I16_200
{Arabidopsis thaliana}, partial (81%)
Length = 1581
Score = 196 bits (497), Expect = 2e-50
Identities = 113/295 (38%), Positives = 177/295 (59%), Gaps = 13/295 (4%)
Frame = +2
Query: 70 VKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVASISRTSHMN 129
+K+ ++F+ +G G V+K L DG VAVK I+ + EF +EV++I+ H+N
Sbjct: 407 LKKQXDNFQAIIGKGSSASVFKGILNDGTSVAVKRIHGEERGEREFRSEVSAIASVQHVN 586
Query: 130 IVSLLGYCYEANK-RALIYEFMPKGSLDKFIYK-SGFPDAVCDC-DSNTLFQIAIGIARG 186
+V L GYC R L+YEF+P GSLD +I+ C C N +++AI +A+
Sbjct: 587 LVRLFGYCNSPTPPRYLVYEFIPNGSLDCWIFPVKETRTRRCGCLPWNLRYKVAIDVAKA 766
Query: 187 LEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYM 246
L YLH C S +LHLD+KP+NILLDEN+ +SDFGL+K+ ++S V + RGT GY+
Sbjct: 767 LSYLHHDCRSTVLHLDVKPENILLDENYKALVSDFGLSKLVGKDESQV-LTTIRGTRGYL 943
Query: 247 APEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQ-----TGGSCTSEMYFPDWIYKDL 301
APE G+S K+D+YS+GM++LE++GGR+N + +FP + + L
Sbjct: 944 APEWLLER--GISEKTDIYSFGMVLLEIVGGRRNVSKVEDPRDNTKKKWQFFPKIVNEKL 1117
Query: 302 EQGN--DLLNSLTI---SEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQG 351
+G ++++ + +EN+ VK++ ++LWCIQ P RP M +V++ML+G
Sbjct: 1118REGKLMEIVDQRVVDFGGVDENE-VKRLVFIALWCIQEKPRLRPSMVEVVDMLEG 1279
>TC93505 weakly similar to GP|15408735|dbj|BAB64138. putative receptor
serine/threonine kinase {Oryza sativa (japonica
cultivar-group)}, partial (10%)
Length = 799
Score = 134 bits (337), Expect(2) = 4e-48
Identities = 66/113 (58%), Positives = 82/113 (72%), Gaps = 1/113 (0%)
Frame = +1
Query: 122 ISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIY-KSGFPDAVCDCDSNTLFQIA 180
I R H+N+V +LG+C E +KRALIYEFM GSLDKFI+ K G + C + + I+
Sbjct: 460 IGRIHHVNVVKILGFCIEGSKRALIYEFMSNGSLDKFIFSKEGSKELCC----SQIHDIS 627
Query: 181 IGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSI 233
IG+ARG+ YLH GC +ILH DIKP NILLDENF PKISDFGLAK+ + +SI
Sbjct: 628 IGVARGIAYLHHGCEMKILHFDIKPHNILLDENFIPKISDFGLAKLYPIXNSI 786
Score = 75.5 bits (184), Expect(2) = 4e-48
Identities = 40/107 (37%), Positives = 65/107 (60%)
Frame = +3
Query: 13 FGVTMFFIIFISCYFKKGIRRPQMTIFRKRRKHVDNNVEVFMQSYNLSIARRYSYAEVKR 72
FG+T F + I +K +RKR + +E+++Q NL + YSY E+K+
Sbjct: 174 FGITFFIALLI---YK----------WRKRHWSMYECIEIYLQQNNL-MPIGYSYREIKK 311
Query: 73 ITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEV 119
+ F+DKLG GG+G V++ + G VA+K++ +SKGNG++FI+EV
Sbjct: 312 MARGFKDKLGEGGFGTVFQGNQRSGPCVAIKMLGKSKGNGQDFISEV 452
>TC88451 similar to GP|836954|gb|AAC23542.1|| receptor protein kinase
{Ipomoea trifida}, partial (28%)
Length = 1276
Score = 184 bits (468), Expect = 5e-47
Identities = 134/403 (33%), Positives = 201/403 (49%), Gaps = 29/403 (7%)
Frame = +3
Query: 28 KKGIRRPQMTIFRKRRKHVD-------NNVEVFMQSYNLSIARRYSYAEVKRITNSF--R 78
KK + P RK K D ++E + +++ + +++ + T F
Sbjct: 69 KKNMHXPDKKSKRKEGKSNDLVESYDIKDLEDDFKGHDIKV---FNFTSILEATMEFSPE 239
Query: 79 DKLGHGGYGVVYKASLTDGRQVAVKVINESKGNG-EEFINEVASISRTSHMNIVSLLGYC 137
+KLG GGYG VYK L G+++AVK ++++ G G EF NE+ I H N+V LLG C
Sbjct: 240 NKLGQGGYGPVYKGILATGQEIAVKRLSKTSGQGIVEFKNELLLICELQHKNLVQLLGCC 419
Query: 138 YEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTL------FQIAIGIARGLEYLH 191
+R LIYE+MP SLD +++ DC L F I GIA+GL YLH
Sbjct: 420 IHEEERILIYEYMPNKSLDFYLF---------DCTKKKLLDWKKRFNIIEGIAQGLLYLH 572
Query: 192 QGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVF 251
+ +I+H D+K NILLDEN PKI+DFG+A++ +S+V+ GT GYM+PE
Sbjct: 573 KYSRLKIIHRDLKASNILLDENMNPKIADFGMARMFTQQESVVNTNRIVGTYGYMSPEYA 752
Query: 252 SRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPD---------W-IYKD- 300
G S KSDVYS+G+L+LE++ G KN + Y D W ++ D
Sbjct: 753 ME--GVCSTKSDVYSFGVLLLEIVCGIKN--------NSFYDVDRPLNLIGHAWELWNDG 902
Query: 301 --LEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTF 358
L+ + LN + +E VK+ V L C++ DRP M++VI +L
Sbjct: 903 EYLKLMDPTLNDTFVPDE----VKRCIHVGLLCVEQYANDRPTMSEVISVLTNKYVLTNL 1070
Query: 359 PPKPVLFSPKRPPLQLSNMSSSDWQETNSITTETELEEEPIEG 401
P KP + + ++S Q+T++ +T T +EG
Sbjct: 1071PRKPAFYVRRE---IFEGETTSKGQDTDTYSTTTISTSFEVEG 1190
>BI272251 similar to GP|13377503|gb TAK33 {Triticum aestivum}, partial (18%)
Length = 582
Score = 184 bits (467), Expect = 6e-47
Identities = 88/179 (49%), Positives = 126/179 (70%)
Frame = +3
Query: 39 FRKRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGR 98
+RK+ +D+ VE F+QS+N + YSY+++K +T F+ KLG GGYG VY+
Sbjct: 48 WRKKHLSIDDMVEDFIQSHNNFMPIXYSYSQIKTMTKHFKHKLGEGGYGSVYEGVSRSKH 227
Query: 99 QVAVKVINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKF 158
+VAVKV+ +S+ NG++FINEVA+I R H+N+V L+G+C E K+AL+YEFMP GSLDK
Sbjct: 228 KVAVKVLTKSQTNGQDFINEVATIGRIRHVNVVQLVGFCAERTKQALVYEFMPNGSLDKH 407
Query: 159 IYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPK 217
++ S + ++ I++GI+RG++YLHQGC +I+H DIKP NILLDENF PK
Sbjct: 408 MF-SHEQGHLSSLSYEKIYDISLGISRGIQYLHQGCDMQIIHFDIKPHNILLDENFDPK 581
>TC78270 similar to GP|16649103|gb|AAL24403.1 Unknown protein {Arabidopsis
thaliana}, partial (80%)
Length = 1463
Score = 181 bits (460), Expect = 4e-46
Identities = 122/345 (35%), Positives = 186/345 (53%), Gaps = 20/345 (5%)
Frame = +3
Query: 65 YSYAEVKRITNSFR--DKLGHGGYGVVYKASLTDGRQVAVKVIN-ESKGNGEEFINEVAS 121
Y+Y E+K +++F +K+G GG+G VYK L G+ A+KV++ ESK +EF+ E+
Sbjct: 273 YTYKELKIASDNFSPANKIGEGGFGSVYKGVLKGGKLAAIKVLSTESKQGVKEFLTEINV 452
Query: 122 ISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAI 181
IS H N+V L G C E + R L+Y ++ SL + + G + D + +I +
Sbjct: 453 ISEIKHENLVILYGCCVEGDHRILVYNYLENNSLSQTLLAGGHSNIYFDWQTRR--RICL 626
Query: 182 GIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRG 241
G+ARGL +LH+ I+H DIK NILLD++ PKISDFGLAK+ + VS G
Sbjct: 627 GVARGLAFLHEEVLPHIVHRDIKASNILLDKDLTPKISDFGLAKLIPSYMTHVS-TRVAG 803
Query: 242 TIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYF-PDWIYKD 300
TIGY+APE R G ++ K+D+YS+G+L++E++ GR N T + W +
Sbjct: 804 TIGYLAPEYAIR--GQLTRKADIYSFGVLLVEIVSGRSNTNTRLPIADQYILETTWQLYE 977
Query: 301 LEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLS-SVTFP 359
++ L++ E + + KI ++L C Q P RP M+ V++ML G + + T
Sbjct: 978 RKELAQLVDISLNGEFDAEEACKILKIALLCTQDTPKLRPTMSSVVKMLTGEMDINETKI 1157
Query: 360 PKPVLFS---------PKR------PPLQLSNMSSSDWQETNSIT 389
KP L S PK+ P + SSSD Q T +I+
Sbjct: 1158TKPGLISDVMDLKIREPKKNINMGTAPSSYNASSSSDSQGTTTIS 1292
>TC88392 similar to GP|11275527|dbj|BAB18292. putative receptor-like protein
kinase {Oryza sativa (japonica cultivar-group)}, partial
(61%)
Length = 1017
Score = 181 bits (459), Expect = 5e-46
Identities = 112/278 (40%), Positives = 160/278 (57%), Gaps = 6/278 (2%)
Frame = +1
Query: 74 TNSFRD--KLGHGGYGVVYKASLTDGRQVAVKVIN-ESKGNGEEFINEVASISRTSHMNI 130
TN F + +LG GG+G V+K + +G +VA+K ++ ES+ EF NEV + R H N+
Sbjct: 208 TNFFSELNQLGRGGFGPVFKGLMPNGEEVAIKKLSMESRQGIREFTNEVRLLLRIQHKNL 387
Query: 131 VSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYL 190
V+LLG C E ++ L+YE++P SLD F++ D D T F+I GIARGL YL
Sbjct: 388 VTLLGCCAEGPEKMLVYEYLPNKSLDHFLF-----DKKRSLDWMTRFRIVTGIARGLLYL 552
Query: 191 HQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEV 250
H+ RI+H DIK NILLDE PKISDFGLA++ D+ V T GYMAPE
Sbjct: 553 HEEAPERIIHRDIKASNILLDEKLNPKISDFGLARLFPGEDTHVQTFRISRTHGYMAPEY 732
Query: 251 FSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTG-GSCTSEMYFPDWIYKDLEQGNDLLN 309
R G +S K+DV+SYG+L+LE++ GRKN+ + +++ W L QG +++
Sbjct: 733 ALR--GYLSVKTDVFSYGVLVLEIVSGRKNHDLKLDAEKADLLSYAW---KLYQGRKIMD 897
Query: 310 SL--TISEEENDMVKKITMVSLWCIQTNPLDRPPMNKV 345
+ I + D + L C Q + ++RP MN V
Sbjct: 898 LIDQNIGKYNGDEAAMCIQLGLLCCQASLVERPDMNSV 1011
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,142,175
Number of Sequences: 36976
Number of extensions: 188649
Number of successful extensions: 2543
Number of sequences better than 10.0: 682
Number of HSP's better than 10.0 without gapping: 1832
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1867
length of query: 459
length of database: 9,014,727
effective HSP length: 99
effective length of query: 360
effective length of database: 5,354,103
effective search space: 1927477080
effective search space used: 1927477080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147875.5


