

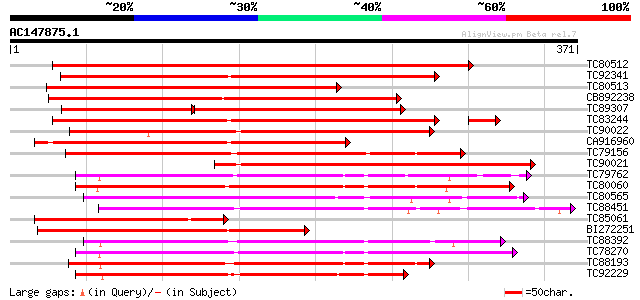
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147875.1 + phase: 0
(371 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC80512 weakly similar to PIR|A96693|A96693 probable receptor se... 488 e-138
TC92341 similar to GP|21741394|emb|CAD39676. OSJNBb0051N19.7 {Or... 340 5e-94
TC80513 weakly similar to GP|9755449|gb|AAF98210.1| Unknown prot... 339 9e-94
CB892238 weakly similar to GP|21741394|em OSJNBb0051N19.7 {Oryza... 315 2e-86
TC89307 similar to GP|21741394|emb|CAD39676. OSJNBb0051N19.7 {Or... 204 2e-79
TC83244 weakly similar to GP|11034602|dbj|BAB17126. putative rec... 270 3e-76
TC90022 similar to GP|14587271|dbj|BAB61189. putative receptor-l... 266 9e-72
CA916960 weakly similar to GP|14090203|db putative receptor kina... 225 2e-59
TC79156 weakly similar to GP|11034603|dbj|BAB17127. putative rec... 210 8e-55
TC90021 weakly similar to GP|13377502|gb|AAK20740.1 LRK33 {Triti... 209 2e-54
TC79762 similar to PIR|H96731|H96731 hypothetical protein F5A18.... 199 1e-51
TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.... 197 4e-51
TC80565 weakly similar to GP|14335072|gb|AAK59800.1 AT5g20050/F2... 193 8e-50
TC88451 similar to GP|836954|gb|AAC23542.1|| receptor protein ki... 191 4e-49
TC85061 similar to GP|9755449|gb|AAF98210.1| Unknown protein {Ar... 190 9e-49
BI272251 similar to GP|13377503|gb TAK33 {Triticum aestivum}, pa... 189 1e-48
TC88392 similar to GP|11275527|dbj|BAB18292. putative receptor-l... 189 2e-48
TC78270 similar to GP|16649103|gb|AAL24403.1 Unknown protein {Ar... 187 7e-48
TC88193 similar to PIR|G96602|G96602 probable receptor protein k... 185 3e-47
TC92229 similar to PIR|B86369|B86369 hypothetical protein AAC980... 182 1e-46
>TC80512 weakly similar to PIR|A96693|A96693 probable receptor
serine/threonine kinase PR5K T4O24.7 [imported] -
Arabidopsis thaliana, partial (35%)
Length = 1077
Score = 488 bits (1256), Expect = e-138
Identities = 240/275 (87%), Positives = 256/275 (92%)
Frame = +1
Query: 29 VEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETK 88
VEVFM+S+NLS+ RRYSYAEVK ITN FR+KLG GGYGVVYKASL + RQVAVKVI+E+K
Sbjct: 253 VEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESK 432
Query: 89 GNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAIC 148
GNGEEFINEVASISRTSHMNIVSLLGYCYE NKRALIYEFMPKGSL+KFIYKS FP+A+C
Sbjct: 433 GNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLNKFIYKSGFPDAVC 612
Query: 149 DFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQR 208
DFD NTLF+IAIGIA+GLEYLHQGCSSRILHLDIKPQNILLDE+FCPKISDFGLAKICQ
Sbjct: 613 DFDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQM 792
Query: 209 KDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTS 268
DSIVSI G RGTIGYMAPE+FSRAFGGVSY+SDVYSYGMLILEMIGGRKNY TGGSCTS
Sbjct: 793 NDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTS 972
Query: 269 EMYFPDWIYKDLEQGNTLLNCSTISEEENDMIRKI 303
EMYFPDWIYKDLEQGN L N TISEEENDM +KI
Sbjct: 973 EMYFPDWIYKDLEQGNDLXNSLTISEEENDMXKKI 1077
>TC92341 similar to GP|21741394|emb|CAD39676. OSJNBb0051N19.7 {Oryza
sativa}, partial (60%)
Length = 779
Score = 340 bits (872), Expect = 5e-94
Identities = 157/248 (63%), Positives = 202/248 (81%)
Frame = +2
Query: 34 RSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGEE 93
+ H + +RY Y+E+K +TN ++KLGQGG+GVVYK L+N VAVK+++ +KGNGEE
Sbjct: 2 KDHGALLQKRYKYSEIKNMTNSLKDKLGQGGFGVVYKGKLFNGCLVAVKILNVSKGNGEE 181
Query: 94 FINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWN 153
FINEVASISRTSH+N+V+LLG+C+E NK+AL+YEFM GSLDKFIY E I W+
Sbjct: 182 FINEVASISRTSHVNVVTLLGFCFEGNKKALVYEFMSNGSLDKFIYNKEL-ETIASLSWD 358
Query: 154 TLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIV 213
L++IA GIA+GLEYLH GC++RILH DIKP NILLD++ CPKISDFGLAK+C RK+SIV
Sbjct: 359 KLYKIAKGIARGLEYLHGGCTTRILHFDIKPHNILLDDNLCPKISDFGLAKLCLRKESIV 538
Query: 214 SILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFP 273
S+ RGT+GY+APE+++R FGGVS++SDVYSYGM++LEM+GGRKN + S TSE+YFP
Sbjct: 539 SMSDQRGTMGYVAPEVWNRHFGGVSHKSDVYSYGMILLEMVGGRKNINADASRTSEIYFP 718
Query: 274 DWIYKDLE 281
W+YK LE
Sbjct: 719 HWVYKRLE 742
>TC80513 weakly similar to GP|9755449|gb|AAF98210.1| Unknown protein
{Arabidopsis thaliana}, partial (14%)
Length = 824
Score = 339 bits (870), Expect = 9e-94
Identities = 164/193 (84%), Positives = 181/193 (92%)
Frame = +2
Query: 25 VDNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVI 84
VD+NVEVFM+S+NLS+ RRYSY EVK ITN FR+KLG GGYGVVYKASL + RQVAVKVI
Sbjct: 233 VDSNVEVFMQSYNLSIARRYSYTEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVI 412
Query: 85 SETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFP 144
+E+KGNGEEFINEVASISRTSH+NIVSLLG+CYE NKRALIYE+MPKGSLDKFIYKS FP
Sbjct: 413 NESKGNGEEFINEVASISRTSHLNIVSLLGFCYEVNKRALIYEYMPKGSLDKFIYKSGFP 592
Query: 145 NAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAK 204
+A+CDFDWNTLF++AIGIA+GLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAK
Sbjct: 593 DAVCDFDWNTLFQVAIGIARGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAK 772
Query: 205 ICQRKDSIVSILG 217
ICQRKDSI +G
Sbjct: 773 ICQRKDSICVNIG 811
>CB892238 weakly similar to GP|21741394|em OSJNBb0051N19.7 {Oryza sativa},
partial (61%)
Length = 726
Score = 315 bits (806), Expect = 2e-86
Identities = 141/231 (61%), Positives = 191/231 (82%)
Frame = +3
Query: 26 DNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVIS 85
D N+E F++ H + +RY Y+E+K +T+ F+ KLGQGG+GVVYK L+N VA+K+++
Sbjct: 36 DTNIEAFLKDHGALLQKRYKYSEIKKMTDSFKVKLGQGGFGVVYKGKLFNGCHVAIKILN 215
Query: 86 ETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPN 145
+KGNGEEFINEV+SI+RTSH+N+V+LLG+C+E K+ALIYEFM GSLDKFIY ++ P
Sbjct: 216 SSKGNGEEFINEVSSITRTSHVNVVTLLGFCFEGTKKALIYEFMSNGSLDKFIY-NKGPE 392
Query: 146 AICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKI 205
I W L++IA GIA+GLEYLH+GC++RILH DIKP NILLDE+ CPKISDFGLAK+
Sbjct: 393 TIASLSWENLYQIAKGIARGLEYLHRGCTTRILHFDIKPHNILLDENLCPKISDFGLAKL 572
Query: 206 CQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGG 256
C +++SI+S+ RGT+GY+APE+++R FGGVS++SDVYSYGM++LEM+GG
Sbjct: 573 CPKQESIISMSDQRGTMGYVAPEVWNRHFGGVSHKSDVYSYGMMLLEMVGG 725
>TC89307 similar to GP|21741394|emb|CAD39676. OSJNBb0051N19.7 {Oryza
sativa}, partial (58%)
Length = 814
Score = 204 bits (519), Expect(2) = 2e-79
Identities = 90/139 (64%), Positives = 122/139 (87%), Gaps = 1/139 (0%)
Frame = +2
Query: 122 RALIYEFMPKGSLDKFIYKSEFPNAI-CDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHL 180
+ALIY++MP GSL+KFIY+++ P + T++ IAIG+A+GLEYLH+GC+++ILH
Sbjct: 398 KALIYDYMPNGSLEKFIYENKDPLKLNLQLSCKTVYNIAIGVARGLEYLHKGCNTKILHF 577
Query: 181 DIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYR 240
DIKP NILLD+DFCPK+SDFGLAK+C RK+SI+S+LGARGT GY+APE+FSR FGGVS++
Sbjct: 578 DIKPHNILLDDDFCPKVSDFGLAKVCPRKESIISLLGARGTAGYIAPEVFSRNFGGVSHK 757
Query: 241 SDVYSYGMLILEMIGGRKN 259
SDVY+YGM++LEM+GG++N
Sbjct: 758 SDVYNYGMMVLEMVGGKQN 814
Score = 110 bits (274), Expect(2) = 2e-79
Identities = 55/88 (62%), Positives = 67/88 (75%)
Frame = +3
Query: 35 SHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGEEF 94
+H +RY+YAE+K TN F+ KLGQGGYG VYK L + VAVKV+SE++GNGEEF
Sbjct: 138 NHGHLAAKRYTYAEIKKATNSFKYKLGQGGYGSVYKGKLQDGSLVAVKVLSESEGNGEEF 317
Query: 95 INEVASISRTSHMNIVSLLGYCYEENKR 122
INEVASIS TSH+NIV LLG+ E +KR
Sbjct: 318 INEVASISVTSHVNIVGLLGFYLEGSKR 401
>TC83244 weakly similar to GP|11034602|dbj|BAB17126. putative receptor
kinase {Oryza sativa (japonica cultivar-group)}, partial
(36%)
Length = 1366
Score = 270 bits (690), Expect(2) = 3e-76
Identities = 126/253 (49%), Positives = 178/253 (69%)
Frame = +1
Query: 29 VEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETK 88
+E F+ + P R++YA++K ITN F+E LG+G +G V+K L VAVKV++ET+
Sbjct: 496 IEKFLEDYRALKPTRFTYADIKRITNGFKESLGEGAHGSVFKGMLSQEILVAVKVLNETQ 675
Query: 89 GNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAIC 148
G+G +FINEV ++ + H+N+V LLG+C + RAL+Y+F P GSL F+ E N
Sbjct: 676 GDGNDFINEVGTMGKIHHVNVVRLLGFCADGFHRALVYDFFPNGSLQNFLAPPE--NKEV 849
Query: 149 DFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQR 208
L RIA+G+A+G+EYLH GC RILH DI P N+L+D++ PKI+DFGLAK+C +
Sbjct: 850 FLGREKLQRIALGVARGIEYLHIGCDHRILHFDINPHNVLIDDNLSPKITDFGLAKLCPK 1029
Query: 209 KDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTS 268
S VS+ ARGT+GY+APE+FSR FG VSY+SD+YSYGML+LEM+GGRKN + T
Sbjct: 1030NQSTVSMTAARGTLGYIAPEVFSRNFGNVSYKSDIYSYGMLLLEMVGGRKNTNQSAKETF 1209
Query: 269 EMYFPDWIYKDLE 281
++ +P+WI+ +E
Sbjct: 1210QVLYPEWIHNLIE 1248
Score = 33.5 bits (75), Expect(2) = 3e-76
Identities = 13/21 (61%), Positives = 15/21 (70%)
Frame = +2
Query: 301 RKITLVSLWCIQTKPSDRPPM 321
+K+ LV LWCIQ P DRP M
Sbjct: 1301 KKLALVGLWCIQWNPVDRPSM 1363
>TC90022 similar to GP|14587271|dbj|BAB61189. putative receptor-like kinase
{Oryza sativa (japonica cultivar-group)}, partial (31%)
Length = 773
Score = 266 bits (680), Expect = 9e-72
Identities = 125/241 (51%), Positives = 179/241 (73%), Gaps = 2/241 (0%)
Frame = +1
Query: 40 MPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKG--NGEEFINE 97
MP RYSY E+K +TN F++KLG+GGYG VYK L + VA+K++ + KG NG++FINE
Sbjct: 4 MPIRYSYTEIKKMTNGFKDKLGEGGYGKVYKGKLRSGPSVAIKMLGKHKGKGNGQDFINE 183
Query: 98 VASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFR 157
VA+I R H N+V L+G+C E +KRAL+Y+FMP GSLDK+I E ++ + ++
Sbjct: 184 VATIGRIHHTNVVRLIGFCVEGSKRALVYDFMPNGSLDKYISSREDHISLT---YKQIYE 354
Query: 158 IAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILG 217
I++ +A+G+ YLHQGC +ILH DIKP NILLD+DF K+SDFGLAK+ ++SIV++
Sbjct: 355 ISLAVARGIAYLHQGCDMQILHFDIKPHNILLDQDFIAKVSDFGLAKLYPIENSIVTMTA 534
Query: 218 ARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIY 277
ARGTIGYMAPE+F + G VSY++DVYS+GML++E+ R+N ++ +S+++FP WIY
Sbjct: 535 ARGTIGYMAPELFYKNIGKVSYKADVYSFGMLLMEIANRRRNLNSNADDSSQIFFPYWIY 714
Query: 278 K 278
+
Sbjct: 715 Q 717
>CA916960 weakly similar to GP|14090203|db putative receptor kinase {Oryza
sativa (japonica cultivar-group)}, partial (27%)
Length = 651
Score = 225 bits (574), Expect = 2e-59
Identities = 111/207 (53%), Positives = 143/207 (68%)
Frame = +2
Query: 17 ILRERTELVDNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNS 76
ILR+ + + +E F+ + P RYSY E+K ITN F + LGQG YG VYK S+
Sbjct: 44 ILRKEKQAI---IEKFLEDYRALKPTRYSYVEIKRITNNFGDMLGQGAYGTVYKGSISKE 214
Query: 77 RQVAVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDK 136
VAVK+++ ++GNG++F+NEV ++ R H+NIV L+G+C + KRALIYEF+P GSL K
Sbjct: 215 FSVAVKILNVSQGNGQDFLNEVGTMGRIHHVNIVRLIGFCADGFKRALIYEFLPNGSLQK 394
Query: 137 FIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPK 196
FI + N W L IA+GIAKG+EYLHQGC RILH DIKPQN+LLD +F PK
Sbjct: 395 FINSPD--NKKNFLGWKKLHEIALGIAKGIEYLHQGCDQRILHFDIKPQNVLLDHNFIPK 568
Query: 197 ISDFGLAKICQRKDSIVSILGARGTIG 223
I DFGLAK+C R SIVS+ ARGT+G
Sbjct: 569 ICDFGLAKLCSRDQSIVSMTAARGTLG 649
>TC79156 weakly similar to GP|11034603|dbj|BAB17127. putative receptor
kinase {Oryza sativa (japonica cultivar-group)}, partial
(18%)
Length = 1246
Score = 210 bits (534), Expect = 8e-55
Identities = 113/263 (42%), Positives = 166/263 (62%), Gaps = 1/263 (0%)
Frame = +1
Query: 37 NLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVIS-ETKGNGEEFI 95
N P R++ ++ IT + LG G +GVV+K L N VAVKV++ G E+F
Sbjct: 469 NREKPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFK 648
Query: 96 NEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTL 155
EV +I RT H+N+V L G+C+ +KRAL+YE++ GSLDK+I+ S+ N DFD+ L
Sbjct: 649 AEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRN---DFDFQKL 819
Query: 156 FRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSI 215
+IAIG AKG+ YLH+ C RI+H DIKP+N+LLD PKI+DFGLAK+ R+ +I
Sbjct: 820 HKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELN 999
Query: 216 LGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDW 275
RGT GY APE++ V+Y+ DVYS+G+L+ E++G R+++D+ S S+ +FP W
Sbjct: 1000THFRGTRGYAAPEMWKPY--PVTYKCDVYSFGILLFEIVGRRRHFDSSYS-ESQQWFPRW 1170
Query: 276 IYKDLEQGNTLLNCSTISEEEND 298
++ E ++ + EE D
Sbjct: 1171TWEMFENNELVVMLALCEIEEKD 1239
>TC90021 weakly similar to GP|13377502|gb|AAK20740.1 LRK33 {Triticum
aestivum}, partial (23%)
Length = 736
Score = 209 bits (531), Expect = 2e-54
Identities = 95/210 (45%), Positives = 146/210 (69%)
Frame = +3
Query: 135 DKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFC 194
D++I+ P ++ + ++ I++G+A+G+ YLHQGC +ILH DIKP NILLD+DF
Sbjct: 3 DRYIFL*RRPISLT---YKQMYEISLGVARGIAYLHQGCDMQILHFDIKPHNILLDQDFI 173
Query: 195 PKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMI 254
K+SDFGLAK+ +SIV++ ARGTIGYMAPE+F + G VSY++DVYS+GML++E+
Sbjct: 174 AKVSDFGLAKLYPVDNSIVTLTAARGTIGYMAPELFYKNIGKVSYKADVYSFGMLLMEIA 353
Query: 255 GGRKNYDTGGSCTSEMYFPDWIYKDLEQGNTLLNCSTISEEENDMIRKITLVSLWCIQTK 314
R+N ++ +S+++FP WIY +L + + ++EE ++K+ +++LWCIQ
Sbjct: 354 NRRRNLNSNADDSSQIFFPYWIYNELMEEREIDISGEATDEEKKNVKKMFIIALWCIQLN 533
Query: 315 PSDRPPMNKVIEMLQGPLSSVSYPPKPVLY 344
P DRP M++VIEML+G + + PPKP Y
Sbjct: 534 PIDRPSMDRVIEMLEGDIEDIEIPPKPSPY 623
>TC79762 similar to PIR|H96731|H96731 hypothetical protein F5A18.8
[imported] - Arabidopsis thaliana, partial (55%)
Length = 1564
Score = 199 bits (506), Expect = 1e-51
Identities = 122/306 (39%), Positives = 174/306 (55%), Gaps = 8/306 (2%)
Frame = +3
Query: 44 YSYAEVKMITNYFR--EKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGE-EFINEVAS 100
+SY + T F KLG+GG+G VYK L + R+VAVK +S+T G+ EF+NE
Sbjct: 348 FSYETLLSATKNFNATHKLGEGGFGPVYKGKLSDGREVAVKKLSQTSNQGKKEFMNEAKL 527
Query: 101 ISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAI 160
++R H N+V+LLGYC ++ L+YE++P SLDKF++K DW F I
Sbjct: 528 LARVQHKNVVNLLGYCVHGTEKILVYEYVPHESLDKFLFKEAEKRE--QLDWKRRFGIIT 701
Query: 161 GIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARG 220
G+AKGL YLH+ + I+H DIK NILLD+ + KI+DFG+A++ S V A G
Sbjct: 702 GVAKGLLYLHEDSHNCIIHRDIKASNILLDDKWTAKIADFGMARLFPEDQSQVKTRVA-G 878
Query: 221 TIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYKDL 280
T GYMAPE G +S ++DV+SYG+L+LE+I G++N + E DW YK
Sbjct: 879 TNGYMAPEYMMH--GRLSVKADVFSYGVLVLELITGQRN-SSFNLXVEEHNLLDWAYKMY 1049
Query: 281 EQGNTL-----LNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSV 335
++G +L ST+ E+ DM ++ L+ CIQ P RP M +++ L S
Sbjct: 1050KKGRSLEIVDSALASTVLTEQVDMCIQLALL---CIQGDPQLRPTMRRIVVKL-----SR 1205
Query: 336 SYPPKP 341
P KP
Sbjct: 1206KSPTKP 1223
>TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.24
[imported] - Arabidopsis thaliana, partial (32%)
Length = 1547
Score = 197 bits (502), Expect = 4e-51
Identities = 114/292 (39%), Positives = 176/292 (60%), Gaps = 5/292 (1%)
Frame = +1
Query: 44 YSYAEVKMITNYF--REKLGQGGYGVVYKASLYNSRQVAVKVIS-ETKGNGEEFINEVAS 100
YS ++K+ TN F + K+G+GG+G VYK L + +AVK +S ++K EF+NE+
Sbjct: 157 YSLRQIKVATNNFDPKNKIGEGGFGPVYKGVLSDGAVIAVKQLSSKSKQGNREFVNEIGM 336
Query: 101 ISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAI 160
IS H N+V L G C E N+ L+YE+M SL + ++ P + DW T +I +
Sbjct: 337 ISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGK--PEQRLNLDWRTRMKICV 510
Query: 161 GIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARG 220
GIA+GL YLH+ +I+H DIK N+LLD++ KISDFGLAK+ + +++ +S A G
Sbjct: 511 GIARGLAYLHEESRLKIVHRDIKATNVLLDKNLNAKISDFGLAKLDEEENTHISTRIA-G 687
Query: 221 TIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYKDL 280
TIGYMAPE R G ++ ++DVYS+G++ LE++ G N + +Y DW Y
Sbjct: 688 TIGYMAPEYAMR--GYLTDKADVYSFGVVALEIVSGMSNTNYRPK-EEFVYLLDWAYVLQ 858
Query: 281 EQGN--TLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQG 330
EQGN L++ + S+ ++ ++ ++L C P+ RPPM+ V+ ML+G
Sbjct: 859 EQGNLLELVDPTLGSKYSSEEAMRMLQLALLCTNPSPTLRPPMSSVVSMLEG 1014
>TC80565 weakly similar to GP|14335072|gb|AAK59800.1 AT5g20050/F28I16_200
{Arabidopsis thaliana}, partial (81%)
Length = 1581
Score = 193 bits (491), Expect = 8e-50
Identities = 114/307 (37%), Positives = 179/307 (58%), Gaps = 16/307 (5%)
Frame = +2
Query: 49 VKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGEEFINEVASISRTSHMN 108
+K + F+ +G+G V+K L + VAVK I + EF +EV++I+ H+N
Sbjct: 407 LKKQXDNFQAIIGKGSSASVFKGILNDGTSVAVKRIHGEERGEREFRSEVSAIASVQHVN 586
Query: 109 IVSLLGYCYEENK-RALIYEFMPKGSLDKFIYK-SEFPNAICD-FDWNTLFRIAIGIAKG 165
+V L GYC R L+YEF+P GSLD +I+ E C WN +++AI +AK
Sbjct: 587 LVRLFGYCNSPTPPRYLVYEFIPNGSLDCWIFPVKETRTRRCGCLPWNLRYKVAIDVAKA 766
Query: 166 LEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYM 225
L YLH C S +LHLD+KP+NILLDE++ +SDFGL+K+ + +S V + RGT GY+
Sbjct: 767 LSYLHHDCRSTVLHLDVKPENILLDENYKALVSDFGLSKLVGKDESQV-LTTIRGTRGYL 943
Query: 226 APE-IFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYD-----TGGSCTSEMYFPDWIYKD 279
APE + R G+S ++D+YS+GM++LE++GGR+N + +FP + +
Sbjct: 944 APEWLLER---GISEKTDIYSFGMVLLEIVGGRRNVSKVEDPRDNTKKKWQFFPKIVNEK 1114
Query: 280 LEQGNTL-------LNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPL 332
L +G + ++ + E E ++++ ++LWCIQ KP RP M +V++ML+G +
Sbjct: 1115LREGKLMEIVDQRVVDFGGVDENE---VKRLVFIALWCIQEKPRLRPSMVEVVDMLEGRV 1285
Query: 333 SSVSYPP 339
V PP
Sbjct: 1286-RVEEPP 1303
>TC88451 similar to GP|836954|gb|AAC23542.1|| receptor protein kinase
{Ipomoea trifida}, partial (28%)
Length = 1276
Score = 191 bits (485), Expect = 4e-49
Identities = 123/324 (37%), Positives = 183/324 (55%), Gaps = 12/324 (3%)
Frame = +3
Query: 59 KLGQGGYGVVYKASLYNSRQVAVKVISETKGNG-EEFINEVASISRTSHMNIVSLLGYCY 117
KLGQGGYG VYK L +++AVK +S+T G G EF NE+ I H N+V LLG C
Sbjct: 243 KLGQGGYGPVYKGILATGQEIAVKRLSKTSGQGIVEFKNELLLICELQHKNLVQLLGCCI 422
Query: 118 EENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRI 177
E +R LIYE+MP SLD +++ + DW F I GIA+GL YLH+ +I
Sbjct: 423 HEEERILIYEYMPNKSLDFYLFDCTKKKLL---DWKKRFNIIEGIAQGLLYLHKYSRLKI 593
Query: 178 LHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGV 237
+H D+K NILLDE+ PKI+DFG+A++ +++S+V+ GT GYM+PE G
Sbjct: 594 IHRDLKASNILLDENMNPKIADFGMARMFTQQESVVNTNRIVGTYGYMSPEYAME--GVC 767
Query: 238 SYRSDVYSYGMLILEMIGGRKN---YDTGGSCTSEMYFPDW-IYKD---LEQGNTLLNCS 290
S +SDVYS+G+L+LE++ G KN YD + W ++ D L+ + LN +
Sbjct: 768 STKSDVYSFGVLLLEIVCGIKNNSFYDVDRPL--NLIGHAWELWNDGEYLKLMDPTLNDT 941
Query: 291 TISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSYPPKPVLYSPERPE 350
+ +E +++ V L C++ +DRP M++VI +L + P KP Y R E
Sbjct: 942 FVPDE----VKRCIHVGLLCVEQYANDRPTMSEVISVLTNKYVLTNLPRKPAFY--VRRE 1103
Query: 351 LQVSDMSS----SDLYETNSVTVS 370
+ + +S +D Y T +++ S
Sbjct: 1104IFEGETTSKGQDTDTYSTTTISTS 1175
>TC85061 similar to GP|9755449|gb|AAF98210.1| Unknown protein {Arabidopsis
thaliana}, partial (8%)
Length = 1013
Score = 190 bits (482), Expect = 9e-49
Identities = 96/128 (75%), Positives = 116/128 (90%), Gaps = 1/128 (0%)
Frame = +1
Query: 17 ILRERTELVDNNVEVFMRSHNLSMP-RRYSYAEVKMITNYFREKLGQGGYGVVYKASLYN 75
ILR+ T+ +DNNVE F++S+NLS+P ++Y YAEVK +TN FR+KLGQGGYGVVYKA+L +
Sbjct: 628 ILRKTTKSIDNNVEDFIQSYNLSVPIKQYRYAEVKKMTNSFRDKLGQGGYGVVYKANLPD 807
Query: 76 SRQVAVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLD 135
RQVAVK+I+E+KGNGE+FINEVASISRTSH+NIVSLLG+CY ENKRALIYEF+PKGSLD
Sbjct: 808 GRQVAVKIINESKGNGEDFINEVASISRTSHVNIVSLLGFCY-ENKRALIYEFLPKGSLD 984
Query: 136 KFIYKSEF 143
KFI KS F
Sbjct: 985 KFILKSGF 1008
>BI272251 similar to GP|13377503|gb TAK33 {Triticum aestivum}, partial (18%)
Length = 582
Score = 189 bits (481), Expect = 1e-48
Identities = 86/178 (48%), Positives = 131/178 (73%)
Frame = +3
Query: 19 RERTELVDNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQ 78
R++ +D+ VE F++SHN MP YSY+++K +T +F+ KLG+GGYG VY+ + +
Sbjct: 51 RKKHLSIDDMVEDFIQSHNNFMPIXYSYSQIKTMTKHFKHKLGEGGYGSVYEGVSRSKHK 230
Query: 79 VAVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFI 138
VAVKV+++++ NG++FINEVA+I R H+N+V L+G+C E K+AL+YEFMP GSLDK +
Sbjct: 231 VAVKVLTKSQTNGQDFINEVATIGRIRHVNVVQLVGFCAERTKQALVYEFMPNGSLDKHM 410
Query: 139 YKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPK 196
+ E + + ++ I++GI++G++YLHQGC +I+H DIKP NILLDE+F PK
Sbjct: 411 FSHE-QGHLSSLSYEKIYDISLGISRGIQYLHQGCDMQIIHFDIKPHNILLDENFDPK 581
>TC88392 similar to GP|11275527|dbj|BAB18292. putative receptor-like protein
kinase {Oryza sativa (japonica cultivar-group)}, partial
(61%)
Length = 1017
Score = 189 bits (479), Expect = 2e-48
Identities = 113/282 (40%), Positives = 164/282 (58%), Gaps = 6/282 (2%)
Frame = +1
Query: 49 VKMITNYFRE--KLGQGGYGVVYKASLYNSRQVAVKVIS-ETKGNGEEFINEVASISRTS 105
+++ TN+F E +LG+GG+G V+K + N +VA+K +S E++ EF NEV + R
Sbjct: 196 LQLATNFFSELNQLGRGGFGPVFKGLMPNGEEVAIKKLSMESRQGIREFTNEVRLLLRIQ 375
Query: 106 HMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKG 165
H N+V+LLG C E ++ L+YE++P SLD F++ + DW T FRI GIA+G
Sbjct: 376 HKNLVTLLGCCAEGPEKMLVYEYLPNKSLDHFLFDKKR-----SLDWMTRFRIVTGIARG 540
Query: 166 LEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYM 225
L YLH+ RI+H DIK NILLDE PKISDFGLA++ +D+ V T GYM
Sbjct: 541 LLYLHEEAPERIIHRDIKASNILLDEKLNPKISDFGLARLFPGEDTHVQTFRISRTHGYM 720
Query: 226 APEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTG-GSCTSEMYFPDWIYKDLEQGN 284
APE R G +S ++DV+SYG+L+LE++ GRKN+D + +++ W L QG
Sbjct: 721 APEYALR--GYLSVKTDVFSYGVLVLEIVSGRKNHDLKLDAEKADLLSYAW---KLYQGR 885
Query: 285 TLLNC--STISEEENDMIRKITLVSLWCIQTKPSDRPPMNKV 324
+++ I + D + L C Q +RP MN V
Sbjct: 886 KIMDLIDQNIGKYNGDEAAMCIQLGLLCCQASLVERPDMNSV 1011
>TC78270 similar to GP|16649103|gb|AAL24403.1 Unknown protein {Arabidopsis
thaliana}, partial (80%)
Length = 1463
Score = 187 bits (474), Expect = 7e-48
Identities = 111/293 (37%), Positives = 169/293 (56%), Gaps = 4/293 (1%)
Frame = +3
Query: 44 YSYAEVKMITNYFR--EKLGQGGYGVVYKASLYNSRQVAVKVIS-ETKGNGEEFINEVAS 100
Y+Y E+K+ ++ F K+G+GG+G VYK L + A+KV+S E+K +EF+ E+
Sbjct: 273 YTYKELKIASDNFSPANKIGEGGFGSVYKGVLKGGKLAAIKVLSTESKQGVKEFLTEINV 452
Query: 101 ISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAI 160
IS H N+V L G C E + R L+Y ++ SL + + N FDW T RI +
Sbjct: 453 ISEIKHENLVILYGCCVEGDHRILVYNYLENNSLSQTLLAGGHSNIY--FDWQTRRRICL 626
Query: 161 GIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARG 220
G+A+GL +LH+ I+H DIK NILLD+D PKISDFGLAK+ + VS A G
Sbjct: 627 GVARGLAFLHEEVLPHIVHRDIKASNILLDKDLTPKISDFGLAKLIPSYMTHVSTRVA-G 803
Query: 221 TIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYF-PDWIYKD 279
TIGY+APE R G ++ ++D+YS+G+L++E++ GR N +T + W +
Sbjct: 804 TIGYLAPEYAIR--GQLTRKADIYSFGVLLVEIVSGRSNTNTRLPIADQYILETTWQLYE 977
Query: 280 LEQGNTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPL 332
++ L++ S E + + KI ++L C Q P RP M+ V++ML G +
Sbjct: 978 RKELAQLVDISLNGEFDAEEACKILKIALLCTQDTPKLRPTMSSVVKMLTGEM 1136
>TC88193 similar to PIR|G96602|G96602 probable receptor protein kinase
F14G9.24 [imported] - Arabidopsis thaliana, partial
(20%)
Length = 1815
Score = 185 bits (469), Expect = 3e-47
Identities = 102/243 (41%), Positives = 154/243 (62%), Gaps = 3/243 (1%)
Frame = +3
Query: 39 SMPRRYSYAEVKMITNYFRE--KLGQGGYGVVYKASLYNSRQVAVKVISETKGNGE-EFI 95
+MP +SY E+K T+ F KLG+GG+G VYK +L + R VAVK +S G+ +FI
Sbjct: 525 TMPNTFSYYELKNATSDFNRDNKLGEGGFGPVYKGTLNDGRFVAVKQLSIGSHQGKSQFI 704
Query: 96 NEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTL 155
E+A+IS H N+V L G C E NKR L+YE++ SLD+ ++ + + +W+T
Sbjct: 705 AEIATISAVQHRNLVKLYGCCIEGNKRLLVYEYLENKSLDQALFGN-----VLFLNWSTR 869
Query: 156 FRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSI 215
+ + +G+A+GL YLH+ RI+H D+K NILLD + PK+SDFGLAK+ K + +S
Sbjct: 870 YDVCMGVARGLTYLHEESRLRIVHRDVKASNILLDSELVPKLSDFGLAKLYDDKKTHIST 1049
Query: 216 LGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDW 275
A GTIGY+APE R G ++ ++DV+S+G++ LE++ GR N D+ +MY DW
Sbjct: 1050RVA-GTIGYLAPEYAMR--GRLTEKADVFSFGVVALELVSGRPNSDSSLE-EDKMYLLDW 1217
Query: 276 IYK 278
++
Sbjct: 1218AWQ 1226
>TC92229 similar to PIR|B86369|B86369 hypothetical protein AAC98010.1
[imported] - Arabidopsis thaliana, partial (44%)
Length = 1347
Score = 182 bits (463), Expect = 1e-46
Identities = 99/221 (44%), Positives = 150/221 (67%), Gaps = 3/221 (1%)
Frame = +2
Query: 44 YSYAEVKMITNYFREK--LGQGGYGVVYKASLYNSRQVAVKVISETKGNGE-EFINEVAS 100
+SY ++ ITN F + +G+GG+G VYKA + + R A+K++ G GE EF EV +
Sbjct: 158 FSYDQILEITNGFSSENVIGEGGFGRVYKALMPDGRVGALKLLKAGSGQGEREFRAEVDT 337
Query: 101 ISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAI 160
ISR H ++VSL+GYC E +R LIYEF+P G+LD+ +++S++ N + DW +IAI
Sbjct: 338 ISRVHHRHLVSLIGYCIAEQQRVLIYEFVPNGNLDQHLHESQW-NVL---DWPKRMKIAI 505
Query: 161 GIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARG 220
G A+GL YLH+GC+ +I+H DIK NILLD+ + +++DFGLA++ ++ VS G
Sbjct: 506 GAARGLAYLHEGCNPKIIHRDIKSSNILLDDSYEAQVADFGLARLTDDTNTHVS-TRVMG 682
Query: 221 TIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYD 261
T GYMAPE + G ++ RSDV+S+G+++LE++ GRK D
Sbjct: 683 TFGYMAPEYATS--GKLTDRSDVFSFGVVLLELVTGRKPVD 799
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,472,976
Number of Sequences: 36976
Number of extensions: 171925
Number of successful extensions: 2283
Number of sequences better than 10.0: 682
Number of HSP's better than 10.0 without gapping: 1555
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1583
length of query: 371
length of database: 9,014,727
effective HSP length: 98
effective length of query: 273
effective length of database: 5,391,079
effective search space: 1471764567
effective search space used: 1471764567
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC147875.1


