

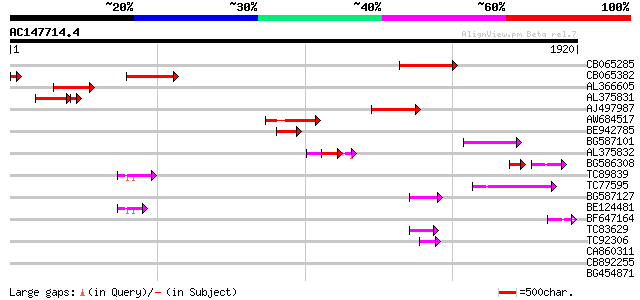
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147714.4 - phase: 0 /pseudo
(1920 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB065285 weakly similar to PIR|A84500|A84 probable retroelement ... 387 e-107
CB065382 similar to PIR|T09671|T096 RPE15 protein - alfalfa (fra... 306 7e-83
AL366605 294 3e-79
AL375831 232 2e-77
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 286 5e-77
AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thalia... 243 5e-64
BE942785 151 2e-36
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 108 2e-23
AL375832 91 3e-18
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 59 3e-17
TC89839 homologue to SP|P23466|CYAA_SACKL Adenylate cyclase (EC ... 84 7e-16
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 74 5e-13
BG587127 weakly similar to PIR|H84506|H84 probable retroelement ... 67 7e-11
BE124481 66 1e-10
BF647164 weakly similar to GP|6691191|gb F7F22.15 {Arabidopsis t... 49 1e-05
TC83629 weakly similar to GP|6692261|gb|AAF24611.1| putative RNa... 47 9e-05
TC92306 45 2e-04
CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product ... 40 0.007
CB892255 39 0.015
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 39 0.015
>CB065285 weakly similar to PIR|A84500|A84 probable retroelement gag/pol
polyprotein [imported] - Arabidopsis thaliana, partial
(2%)
Length = 592
Score = 387 bits (995), Expect = e-107
Identities = 183/197 (92%), Positives = 189/197 (95%)
Frame = -2
Query: 1320 KSKDCEEPLIGEGPDPNSKWGLVFDGAANAYGKGIGAVIVSPQGHHIPFTAQILFECTNN 1379
KSKDCEEPLIGEGPDPNSKWGLVFDGA NAYGKGIGAVIVSPQGH+IPFTA+ILFECTNN
Sbjct: 591 KSKDCEEPLIGEGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGHYIPFTARILFECTNN 412
Query: 1380 MVEYEACIFGIEEAIGMRIKHLDIYGDSALIINQIKGEWETHHAKLIPYRDYARRLLTYF 1439
M EYEACIFGIEEAI MRIKHLDIYGDSAL+INQIKGEWETHHA LIPYRDYARRLLTYF
Sbjct: 411 MAEYEACIFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHANLIPYRDYARRLLTYF 232
Query: 1440 TKVELHHIPRDENQMADALATLSSMFRVNHWNDVPIIKVQRLERPSHVFTIGDVIDQAGE 1499
TKVELHHIPRDENQMADALATLSSMFRVNHWNDVPIIKVQRLERPSHVF IG+VIDQ GE
Sbjct: 231 TKVELHHIPRDENQMADALATLSSMFRVNHWNDVPIIKVQRLERPSHVFAIGNVIDQTGE 52
Query: 1500 NMVDNKPWYYDIKQCLL 1516
N+VD KPWYYDIKQ L+
Sbjct: 51 NVVDYKPWYYDIKQFLV 1
>CB065382 similar to PIR|T09671|T096 RPE15 protein - alfalfa (fragment),
partial (9%)
Length = 624
Score = 306 bits (783), Expect = 7e-83
Identities = 148/179 (82%), Positives = 156/179 (86%)
Frame = -2
Query: 394 PYQKQQLPVQQQQQNQQARPTFPPIPMLYAELLPTLLHRGHCTTRQGKPPPDPLPPRFRS 453
P + PV QQ+Q QQARPTFP IPMLYAE LPTLL RGHCT RQGKPPPDPLPPRFRS
Sbjct: 539 PPRNNHPPVHQQRQ*QQARPTFPLIPMLYAE*LPTLLLRGHCTIRQGKPPPDPLPPRFRS 360
Query: 454 DLKCDFHQGALGHDVEGCYALKYIVKKLIDQGKLTFKNNVPHVLDNPLPNHAAVNMIEVC 513
DLKCDFHQGALGHDVEGCYALK+IVKKLI+QGKLTF+NNVPHVLDNPLPNHAAVNMIEV
Sbjct: 359 DLKCDFHQGALGHDVEGCYALKHIVKKLINQGKLTFENNVPHVLDNPLPNHAAVNMIEVY 180
Query: 514 EEAPRLDVRNVATPLVPLHIKLCKASLFSHDHAKCLGCLRNPLGCYTVQDDIQSLMNDN 572
EEAP LDVRNV TPLVPLHIKLC+ASLF HDHA C NPLGC VQ+DIQSLMN+N
Sbjct: 179 EEAPGLDVRNVTTPLVPLHIKLCQASLFDHDHANCQE*FYNPLGCCVVQNDIQSLMNNN 3
Score = 63.5 bits (153), Expect = 7e-10
Identities = 32/40 (80%), Positives = 35/40 (87%)
Frame = -1
Query: 1 NAQLRTELATLREELAKANDVMTALLAAQEQSATAIPIAT 40
NAQLRTELA++REELAKAND MTALLAAQEQS+T P T
Sbjct: 618 NAQLRTELASMREELAKANDTMTALLAAQEQSSTGSPTTT 499
>AL366605
Length = 422
Score = 294 bits (752), Expect = 3e-79
Identities = 140/140 (100%), Positives = 140/140 (100%)
Frame = -3
Query: 148 HEIKANRGNADSFKTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYKD 207
HEIKANRGNADSFKTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYKD
Sbjct: 420 HEIKANRGNADSFKTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYKD 241
Query: 208 NDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDELAAAFKSHYGFNTRLKPNREFLRSLS 267
NDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDELAAAFKSHYGFNTRLKPNREFLRSLS
Sbjct: 240 NDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDELAAAFKSHYGFNTRLKPNREFLRSLS 61
Query: 268 QKKEESFREYAQRWRGAAAR 287
QKKEESFREYAQRWRGAAAR
Sbjct: 60 QKKEESFREYAQRWRGAAAR 1
>AL375831
Length = 467
Score = 232 bits (591), Expect(2) = 2e-77
Identities = 113/121 (93%), Positives = 116/121 (95%)
Frame = +2
Query: 87 VLIPATNAVSINATLPQTTAAVTEPLVHAIPQGVNINTQHGSIPVTKTMEEMMEELAKEL 146
VLIPATNA SI ATLPQTTAAVTEPLVH +PQG+NINTQH SIPVTKTMEEMMEELAKEL
Sbjct: 2 VLIPATNAASIIATLPQTTAAVTEPLVHTLPQGININTQHRSIPVTKTMEEMMEELAKEL 181
Query: 147 RHEIKANRGNADSFKTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYK 206
RHEI+ANRGNADS KTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYK
Sbjct: 182 RHEIQANRGNADSVKTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYK 361
Query: 207 D 207
D
Sbjct: 362 D 364
Score = 77.8 bits (190), Expect(2) = 2e-77
Identities = 35/36 (97%), Positives = 35/36 (97%)
Frame = +3
Query: 206 KDNDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDE 241
K NDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDE
Sbjct: 360 KTNDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDE 467
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 286 bits (732), Expect = 5e-77
Identities = 125/167 (74%), Positives = 152/167 (90%)
Frame = -2
Query: 1224 KTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAVVTGKIARWQVLLSEYDIVFKT 1283
KTCCALAWAAKRLRHY++NHTTWL+S+MDPIKYIFEK +TG+IARWQ+LLSEYDI +++
Sbjct: 635 KTCCALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPALTGRIARWQMLLSEYDIEYRS 456
Query: 1284 QKAIKGSILADHPAYQPLDDYQPIESDFPNEEIVYLKSKDCEEPLIGEGPDPNSKWGLVF 1343
QKAIKGSILADH A+QPL+DY+PI+ DFP+EEI+YLK KDC+EPL GEGPDP+S WGL+F
Sbjct: 455 QKAIKGSILADHLAHQPLEDYRPIKFDFPDEEIMYLKMKDCDEPLFGEGPDPDSVWGLIF 276
Query: 1344 DGAANAYGKGIGAVIVSPQGHHIPFTAQILFECTNNMVEYEACIFGI 1390
DGA N YG GIGAV+++P+G HIPFTA++ F+CTNN+ EYEACI GI
Sbjct: 275 DGAVNVYGNGIGAVLLTPKGAHIPFTARLRFDCTNNIAEYEACIMGI 135
>AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thaliana chromosome
II BAC F26H6; putative retroelement pol polyprotein,
partial (1%)
Length = 488
Score = 243 bits (620), Expect = 5e-64
Identities = 129/187 (68%), Positives = 142/187 (74%)
Frame = +1
Query: 866 ITFQVMDINASYNCLLGRP*IHDAGAVTSTLHQKLKFAKSGKLVTIHGEEAYLVSQLSSF 925
ITFQVMDINASY+CLLGRP IHDAGAVTSTLHQKLKF K+G
Sbjct: 1 ITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKFVKNG------------------- 123
Query: 926 SCIEAGSAEGTAFQGLTIEGTEPKRDGTAMASLKDAQRAVQEGQAAG*GRLIQLRENKHK 985
SAEGTAFQGL++EG EPK+ G AMASLKDAQ+AVQEGQAA G+LIQL ENK K
Sbjct: 124 ------SAEGTAFQGLSMEGAEPKKVGAAMASLKDAQKAVQEGQAADWGKLIQLCENKRK 285
Query: 986 EGLGFSPTSGVSTGAFYSAGFVNAITEEATGFGPRPVFVIPGGIARDWDAIDIPSIMHVS 1045
EGL FSPTSGVSTG F+SAGFVN + EE F PRP+FVIPGGIA+DWDA+D+PSIMHVS
Sbjct: 286 EGLRFSPTSGVSTGTFHSAGFVNTLAEEVARFVPRPLFVIPGGIAKDWDAVDVPSIMHVS 465
Query: 1046 E*YTLLL 1052
* LLL
Sbjct: 466 X*CVLLL 486
>BE942785
Length = 460
Score = 151 bits (382), Expect = 2e-36
Identities = 76/82 (92%), Positives = 79/82 (95%)
Frame = -2
Query: 905 SGKLVTIHGEEAYLVSQLSSFSCIEAGSAEGTAFQGLTIEGTEPKRDGTAMASLKDAQRA 964
SG LVTIHGEEAYL+SQLSSFSCIEAGSAEGTAFQGLT+EGTEPKRDGTAMASLKDAQRA
Sbjct: 378 SGXLVTIHGEEAYLISQLSSFSCIEAGSAEGTAFQGLTVEGTEPKRDGTAMASLKDAQRA 199
Query: 965 VQEGQAAG*GRLIQLRENKHKE 986
VQE QAAG GRLIQLRENKHK+
Sbjct: 198 VQESQAAGWGRLIQLRENKHKD 133
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 108 bits (271), Expect = 2e-23
Identities = 63/196 (32%), Positives = 96/196 (48%)
Frame = +2
Query: 1538 RFLLDGDILYKRNYDMVLLRCVDEHEAEQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWL 1597
R+L D LYK+ D + +RCV E E ++ H + H K+ +AG++W
Sbjct: 17 RYLWDEPFLYKQCADNIYIRCVAEEEIPGILFHCHGSNYAGHFAVSKTVSKIQQAGFWWP 196
Query: 1598 TMEHDCYQHARKCHKCQIYADKIHVPPHALNVISSPWPFSMVGIDMIGRIEPKASNGHRF 1657
TM D + KC CQ + N I F + GID +G +N ++
Sbjct: 197 TMFKDAHSFISKCDPCQRQGNIS*RNEMPQNFILEVEVFDVWGIDFMGPFPSSYNN--KY 370
Query: 1658 ILVAIDYFTKWVEAASYTNVTKQVVAKFIKNNIICRYGVPSKIITDNGTNLNNNVVQALC 1717
ILVA+DY +KWVEA + VV K K+ I R+GVP +I+D G++ N V + L
Sbjct: 371 ILVAVDYVSKWVEAIASPTNDATVVVKMFKSVIFPRFGVPRVVISDGGSHFINKVFEKLL 550
Query: 1718 EEFKIEHHNSSPYRPQ 1733
++ + H ++ Y PQ
Sbjct: 551 KKNGVRHKVATAYHPQ 598
>AL375832
Length = 535
Score = 91.3 bits (225), Expect = 3e-18
Identities = 71/182 (39%), Positives = 81/182 (44%), Gaps = 12/182 (6%)
Frame = +3
Query: 1006 FVNAITEEATGFGPRPVFVIPGGIARDWDAIDIPSIMHVSE*YTLLLKYLVFSK*QSSRS 1065
FVNAI+EEATG G RP FV PGGIA DWDAIDIPSIMHVSE* LLL VF
Sbjct: 3 FVNAISEEATGSGLRPAFVTPGGIASDWDAIDIPSIMHVSE*CVLLL*RFVFKNNNPLAL 182
Query: 1066 AQSESDIFCKGMFVSALPLINKNFVVSSTTAFLVLFPLEIMVMPEKTKTSVFSY*FQKSA 1125
++ + L+NKN F F LE +K K F F+
Sbjct: 183 PKARVIFSVRACLFRHCRLMNKNCRFFHDCFFFSAFFLEKW*CQKKPKRLYFLINFKNLH 362
Query: 1126 *KTSF*NHPIILRRLNYNKPVGHSNPAFP------------PNFEFPVYEAEDEEGDDIP 1173
K I + + K H P FPVYE + EGDDIP
Sbjct: 363 EKKK-----IQKTKKLFLKSSNHYMQVEP**TR*TQ*FYASSKL*FPVYEPKMREGDDIP 527
Query: 1174 YE 1175
YE
Sbjct: 528 YE 533
Score = 80.9 bits (198), Expect = 4e-15
Identities = 49/73 (67%), Positives = 50/73 (68%)
Frame = +2
Query: 1055 LVFSK*QSSRSAQSESDIFCKGMFVSALPLINKNFVVSSTTAFLVLFPLEIMVMPEKTKT 1114
L F K*QSSRSAQSESDIFCKG+FVSALP S F L MVMPEKT T
Sbjct: 149 LCFQK*QSSRSAQSESDIFCKGVFVSALPFDE*KLSFLSRLLFF*CLFLGKMVMPEKT*T 328
Query: 1115 SVFSY*FQKSA*K 1127
SVFSY*FQK A*K
Sbjct: 329 SVFSY*FQKPA*K 367
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 686
Score = 58.9 bits (141), Expect(2) = 3e-17
Identities = 38/123 (30%), Positives = 68/123 (54%), Gaps = 4/123 (3%)
Frame = -3
Query: 1767 LHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEIPSL---RVIMEAKLSEAEWCQSRYDQL 1823
L +RT R +T +TPFS+ + +EA+ P EV + SL R+ +L+ ++ L
Sbjct: 462 LWSHRTNPRGATKSTPFSMAHRVEAMAPAEVNVTSL*RSRMPQNIELNN----DRLFNAL 295
Query: 1824 NLVEEKRMDAMARGQSYQARMKTAFDKKVHPREFKEGELVLKRRISQQPD-PRGKWTPNY 1882
+EE+R A+ R Q+YQ ++++ ++K V + K G++VL + + GK N+
Sbjct: 294 ETIEERRDQALLRIQNYQHQIESYYNKTVKSQPLKLGDIVLCKVFENTKELNAGKLGTNW 115
Query: 1883 EGS 1885
EG+
Sbjct: 114 EGN 106
Score = 49.7 bits (117), Expect(2) = 3e-17
Identities = 22/52 (42%), Positives = 35/52 (67%)
Frame = -2
Query: 1694 YGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQINGVVEAANKNI 1745
+G+P +I+TDNG++ +N + CE ++I + +SP PQ NG EA+NK I
Sbjct: 685 HGLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKII 530
>TC89839 homologue to SP|P23466|CYAA_SACKL Adenylate cyclase (EC 4.6.1.1)
(ATP pyrophosphate-lyase) (Adenylyl cyclase). [Yeast],
partial (0%)
Length = 665
Score = 83.6 bits (205), Expect = 7e-16
Identities = 59/150 (39%), Positives = 73/150 (48%), Gaps = 18/150 (12%)
Frame = +3
Query: 366 EVGMVSAGAGQSMATVAPINATQMPPSYPYQ-------KQQLPVQQQQQNQQARPTFP-- 416
EV +++ MAT P Q P S PYQ +QQ P Q QNQ + P
Sbjct: 177 EVEKLTSLVSSLMATNDPPLVQQRPQS-PYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQN 353
Query: 417 ---------PIPMLYAELLPTLLHRGHCTTRQGKPPPDPLPPRFRSDLKCDFHQGALGHD 467
PIP+ YA+LLP LL + T P+ LPP +R DL C FHQGA GHD
Sbjct: 354 QVQKASQCDPIPVKYADLLPILLKKNLIQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHD 533
Query: 468 VEGCYALKYIVKKLIDQGKLTFKNNVPHVL 497
E CY LK V+KLI+ +F + VL
Sbjct: 534 TEQCYPLKEEVQKLIENNVWSFDDQDIKVL 623
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 73.9 bits (180), Expect = 5e-13
Identities = 62/291 (21%), Positives = 119/291 (40%), Gaps = 6/291 (2%)
Frame = +2
Query: 1566 QLMHDVHDGTFGTHATGHTMSRKLLRAGYYWLTMEHDCYQHARKCHKCQIYADKIHVPPH 1625
+L+ + HD T H G + +++ ++W + R C C IH+
Sbjct: 176 KLVQESHDSTAAGHP-GRNGTLEIVSRKFFWPGQSQTVRRFVRNCDVC----GGIHIWRQ 340
Query: 1626 ALNVISSPWPF-----SMVGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQ 1680
A P P S + +D I + P G +++ V +D +K V + +
Sbjct: 341 AKRGFLKPLPVPNRLHSDLSMDFITSLPPTRGRGSQYLWVIVDRLSKSVTLEEMDTMEAE 520
Query: 1681 VVAKFIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQINGVVEA 1740
A+ + +G+P I++D G+N + C + S+ Y PQ +G E
Sbjct: 521 ACAQRFLSCHYRFHGMPQSIVSDRGSNWVGRFWREFCRLTGVTQLLSTSYHPQTDGGTER 700
Query: 1741 ANKNIKRIVQKMVTTYRD-WHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEI 1799
N+ I+ +++ V +D W ++LP R SS GATPF + +G V P+
Sbjct: 701 WNQEIQAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSIGATPFFVEHGYH-VDPIPTVE 877
Query: 1800 PSLRVIMEAKLSEAEWCQSRYDQLNLVEEKRMDAMARGQSYQARMKTAFDK 1850
+ V+ E + + + D ++ + + A R ++ + + D+
Sbjct: 878 DTGGVVSEGEAAAQLLVKRMKDVTGFIQAEIVAAQQRSEASANKRRCPADR 1030
>BG587127 weakly similar to PIR|H84506|H84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(13%)
Length = 415
Score = 67.0 bits (162), Expect = 7e-11
Identities = 37/109 (33%), Positives = 56/109 (50%)
Frame = +3
Query: 1355 GAVIVSPQGHHIPFTAQILFECTNNMVEYEACIFGIEEAIGMRIKHLDIYGDSALIINQI 1414
G + SP + + ++ F +NN YEA I G+ A G++I+++ Y DS L+ +Q
Sbjct: 3 GIRLTSPTNEILKQSFRLEFHASNNETNYEALIAGVRLAHGLKIRNIHAYCDSQLVASQF 182
Query: 1415 KGEWETHHAKLIPYRDYARRLLTYFTKVELHHIPRDENQMADALATLSS 1463
GE+E + Y ++L L IPR EN ADALA L+S
Sbjct: 183 SGEYEARDELMDTYLKLVQKLAQKLDYFALTRIPRSENVQADALAALAS 329
>BE124481
Length = 538
Score = 65.9 bits (159), Expect = 1e-10
Identities = 47/120 (39%), Positives = 57/120 (47%), Gaps = 18/120 (15%)
Frame = +3
Query: 366 EVGMVSAGAGQSMATVAPINATQMPPSYPYQ-------KQQLPVQQQQQNQQARPTFP-- 416
EV +++ MAT P Q P S PYQ +QQ P Q QNQ + P
Sbjct: 180 EVEKLTSLVSSLMATNDPPLVQQRPQS-PYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQN 356
Query: 417 ---------PIPMLYAELLPTLLHRGHCTTRQGKPPPDPLPPRFRSDLKCDFHQGALGHD 467
PIP+ YA+LLP LL + T P+ LPP +R DL C FHQGA GHD
Sbjct: 357 QVQKASQCDPIPVKYADLLPILLKKNLIQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHD 536
>BF647164 weakly similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana},
partial (6%)
Length = 469
Score = 49.3 bits (116), Expect = 1e-05
Identities = 35/101 (34%), Positives = 52/101 (50%), Gaps = 3/101 (2%)
Frame = +1
Query: 1822 QLNLVEEKRMDAMARGQSYQARMKTAFDKKVHPREFKEGELVL--KRRISQQPDPRGKWT 1879
+LN +EE R+DA + Y+ R K D+++ REF+EGELVL R+ P GK
Sbjct: 103 ELNELEELRLDAYENAKIYKERTKKWHDRRIIRREFREGELVLLFNSRLKLFP---GKLR 273
Query: 1880 PNYEGSYVVKKAFSGGAL-ILTHMDGVELPNPVNADLVKKY 1919
++ G + VK GA+ + + G P VN +K Y
Sbjct: 274 SHWSGPFQVKNVMPSGAVEVWSESTG---PFTVNGQRLKHY 387
>TC83629 weakly similar to GP|6692261|gb|AAF24611.1| putative RNase H
{Arabidopsis thaliana}, partial (21%)
Length = 1071
Score = 46.6 bits (109), Expect = 9e-05
Identities = 30/100 (30%), Positives = 44/100 (44%)
Frame = +2
Query: 1353 GIGAVIVSPQGHHIPFTAQILFECTNNMVEYEACIFGIEEAIGMRIKHLDIYGDSALIIN 1412
G GA++++ G + Q L T EY A + G++ A K++ GDS L+IN
Sbjct: 602 GAGALLLAEDGSLLYGFRQGLGHQTKESAEYRALLLGLKHASMKGFKYVTAKGDSELVIN 781
Query: 1413 QIKGEWETHHAKLIPYRDYARRLLTYFTKVELHHIPRDEN 1452
QI W+ L A L F + HI R+ N
Sbjct: 782 QILDPWKIKDEHLKKLCAEALELSDNFHSFRIQHISRERN 901
>TC92306
Length = 521
Score = 45.4 bits (106), Expect = 2e-04
Identities = 23/73 (31%), Positives = 35/73 (47%)
Frame = +2
Query: 1387 IFGIEEAIGMRIKHLDIYGDSALIINQIKGEWETHHAKLIPYRDYARRLLTYFTKVELHH 1446
I G+ EA +H+ + GD + Q G W+ ++ L + A L + F V + H
Sbjct: 2 ILGLNEARNQGYEHVHVRGDFQXVCKQFXGSWKVNNPNLRNLCNXAVELKSNFKSVSVEH 181
Query: 1447 IPRDENQMADALA 1459
+PR N ADA A
Sbjct: 182 VPRGXNXAADAQA 220
>CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product {Drosophila
melanogaster}, partial (20%)
Length = 192
Score = 40.4 bits (93), Expect = 0.007
Identities = 17/36 (47%), Positives = 22/36 (60%)
Frame = +1
Query: 1205 IYYLSKKLTDCETRYTMLEKTCCALAWAAKRLRHYL 1240
I Y S+ LT E YT++E+ C A WA + RHYL
Sbjct: 64 IAYASRLLTAAERNYTVVERECLAAIWAIRNFRHYL 171
>CB892255
Length = 853
Score = 39.3 bits (90), Expect = 0.015
Identities = 29/98 (29%), Positives = 47/98 (47%)
Frame = +1
Query: 1786 VYGMEAVLPLEVEIPSLRVIMEAKLSEAEWCQSRYDQLNLVEEKRMDAMARGQSYQARMK 1845
V+ ++A+ ++V++ S+ V +AK+ C R ++RG S +
Sbjct: 31 VFNVKALCHMKVKVLSMEVPKKAKID*T*KCYCRVP---------WTVISRG*SKYST-- 177
Query: 1846 TAFDKKVHPREFKEGELVLKRRISQQPDPRGKWTPNYE 1883
KK PRE +E VLK + P+ RGKW PNY+
Sbjct: 178 ----KKARPRECQESGFVLKNML---PNSRGKWMPNYK 270
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 39.3 bits (90), Expect = 0.015
Identities = 22/65 (33%), Positives = 32/65 (48%), Gaps = 1/65 (1%)
Frame = +2
Query: 1727 SSPYRPQINGVVEAANKNIKRIVQ-KMVTTYRDWHEMLPYALHGYRTTVRSSTGATPFSL 1785
SS Y P +G EA NK + ++ M T W + P+A + Y T+ S TPF
Sbjct: 83 SSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEYWYNTSYNISAAMTPFKA 262
Query: 1786 VYGME 1790
+YG +
Sbjct: 263 LYGRD 277
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.136 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 57,360,519
Number of Sequences: 36976
Number of extensions: 847284
Number of successful extensions: 4608
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 4411
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4593
length of query: 1920
length of database: 9,014,727
effective HSP length: 110
effective length of query: 1810
effective length of database: 4,947,367
effective search space: 8954734270
effective search space used: 8954734270
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC147714.4


