

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
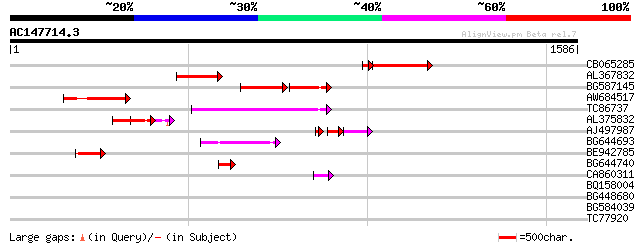
Query= AC147714.3 - phase: 0 /pseudo
(1586 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB065285 weakly similar to PIR|A84500|A84 probable retroelement ... 244 3e-73
AL367832 weakly similar to GP|14091845|gb Putative retroelement ... 244 1e-64
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 150 5e-57
AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thalia... 188 1e-47
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 172 7e-43
AL375832 117 5e-26
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 85 4e-22
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 86 1e-16
BE942785 83 7e-16
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 49 2e-05
CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product ... 47 5e-05
BQ158004 weakly similar to SP|Q8VXP3|TBC2 Tbc2 translation facto... 32 1.5
BG448680 homologue to GP|4249411|gb|A unknown protein {Arabidops... 32 2.0
BG584039 homologue to GP|4249411|gb|A unknown protein {Arabidops... 32 2.0
TC77920 similar to GP|15809948|gb|AAL06901.1 AT3g10210/F14P13_19... 32 2.6
>CB065285 weakly similar to PIR|A84500|A84 probable retroelement gag/pol
polyprotein [imported] - Arabidopsis thaliana, partial
(2%)
Length = 592
Score = 244 bits (622), Expect(2) = 3e-73
Identities = 122/166 (73%), Positives = 134/166 (80%)
Frame = -1
Query: 1016 IW*RNRGNRCIPAGAPYSFYRPDSVRMYKQYGRIRSMYLWDRGSN*HENQTPRHLWRFDT 1075
+W*RN + CIPAGA + FYRPDSVRMYKQYG + SMYLWDRGSN*HENQTPRHLWRF T
Sbjct: 502 LW*RNWSSHCIPAGALHPFYRPDSVRMYKQYG*V*SMYLWDRGSN*HENQTPRHLWRFCT 323
Query: 1076 CHQPDKG*MGDPPCQADSLP*LRETFADIFYKGRIAPYPSG*EPNGRCSSYSILHVSSES 1135
CHQPD+G*MGDPPCQ+DSLP L E FAD+FYK APY S EPNG CS YSILHVSS+
Sbjct: 322 CHQPDQG*MGDPPCQSDSLPRLCEAFADLFYKS*AAPYSS*REPNG*CSGYSILHVSSKP 143
Query: 1136 LEQCANNPSATP*KTFSCVYY*RYDQSG*RECG*R*TMVL*HQVVL 1181
LE CANN SATP*KTF+CV Y D+S +CG* *T+VL HQ VL
Sbjct: 142 LE*CANNQSATP*KTFTCVCYWECDRSDR*KCG*L*TLVLRHQTVL 5
Score = 52.0 bits (123), Expect(2) = 3e-73
Identities = 28/31 (90%), Positives = 30/31 (96%)
Frame = -3
Query: 986 NQKTAKNR*LVKVQIPIASGD*SLMVLSMLI 1016
NQKTAKNR*LVKVQIPIA+G *SLMVLSML+
Sbjct: 590 NQKTAKNR*LVKVQIPIANGV*SLMVLSMLM 498
>AL367832 weakly similar to GP|14091845|gb Putative retroelement {Oryza
sativa}, partial (2%)
Length = 384
Score = 244 bits (624), Expect = 1e-64
Identities = 122/128 (95%), Positives = 123/128 (95%)
Frame = -1
Query: 467 QERKAIQPHQEEIELINLGTEENKREIKVGAALEEGVKRKIFQLL*EYPDIFAWSYEDMP 526
QERKAIQPHQEEIELINLGTEENKREIKVGAALEEGVKRKIFQLL EY DIFA SYEDMP
Sbjct: 384 QERKAIQPHQEEIELINLGTEENKREIKVGAALEEGVKRKIFQLLREYLDIFACSYEDMP 205
Query: 527 GLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVANI 586
GLDPKIVEHRIPTKPECPPVR KLRRTHPDMALKIKSEV KQIDAGFLMT+EYPEWVANI
Sbjct: 204 GLDPKIVEHRIPTKPECPPVR*KLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANI 25
Query: 587 VPVPKKDG 594
VPVPKKDG
Sbjct: 24 VPVPKKDG 1
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 150 bits (379), Expect(2) = 5e-57
Identities = 69/132 (52%), Positives = 100/132 (75%)
Frame = +2
Query: 646 MSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVK 705
M P+D EKT+FIT GT+CYKVMPFGL NAG+TYQR + +F D + +EVY+DDM+VK
Sbjct: 2 MHPDDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVK 181
Query: 706 SKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIR 765
S H+ +L + F+ L +Y ++LNP KCTFGV SG+ LG+IV+Q+GIEV+P ++ AI
Sbjct: 182 SLRATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAIL 361
Query: 766 EMPAPQTEKQVR 777
++P+P+ ++V+
Sbjct: 362 DLPSPKNSREVQ 397
Score = 91.3 bits (225), Expect(2) = 5e-57
Identities = 47/119 (39%), Positives = 72/119 (60%)
Frame = +3
Query: 782 RLNYISRFISHMTATCGPIFKLLRKNQPIVWNDECQGAFDSIKNYLLEPPILVPPMEGKP 841
R+ ++RFIS T C P +KLL N+ VW+++C+ AF+ +K YL PP+L P G
Sbjct: 411 RIAALNRFISRSTDKCLPFYKLLCGNKRFVWDEKCEEAFEQLKQYLTTPPVLSKPEAGDT 590
Query: 842 LIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTDCETRYMMLEKTCCAPAWAAKR 900
L +Y+++ +V VL ++D +K I+Y SK+ TD ETRY LEK A +A++
Sbjct: 591 LSLYIAISSTAVSSVLIREDRGEQK--PIFYTSKRMTDPETRYPTLEKMAFAVITSARK 761
>AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 488
Score = 188 bits (478), Expect = 1e-47
Identities = 103/187 (55%), Positives = 114/187 (60%)
Frame = +3
Query: 150 HIPGHGHQCFL*LSPGKTMNT*CWRSNLYPAPEIEVCKEWEACYSPWRRGIFG*PAVIFL 209
H+ GHG+QCF+*LSPGKT++T* WR NLYP PE+E+C+EW
Sbjct: 3 HVSGHGYQCFV*LSPGKTLDT*RWRGNLYPTPEVEICEEW-------------------- 122
Query: 210 LH*SWIC*RNFLSRFDY*RYGA*EGWGYYGFFKRRSEGCPRRIGCRLGQVNTTPRE*A*R 269
IC* N SR Y R A EGW GFF+ R EGC GCRLGQVN T E*A
Sbjct: 123 -----IC*GNCFSRVVYGRCRAQEGWSCNGFFEGRPEGCSGGTGCRLGQVNPTL*E*AKG 287
Query: 270 RSRVFPNIRCLHKNFP*CWVCQCNY*RGDWIWS*ACVCYTRSHCERLGCHRHSFNHACFR 329
RS++ PNI CLH NF *C VCQ *RG I S*A VC TR HCERLGC S NHAC
Sbjct: 288 RSQILPNIGCLHGNFS*CRVCQYTC*RGGSICS*ALVCDTRRHCERLGCR*CSLNHACVX 467
Query: 330 VMCLVA* 336
MCLVA*
Sbjct: 468 XMCLVA* 488
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 172 bits (437), Expect = 7e-43
Identities = 121/398 (30%), Positives = 196/398 (48%), Gaps = 7/398 (1%)
Frame = +1
Query: 510 LL*EYPDIFAWSYEDMPGLDPKIVEHRIPTKPEC----PPVRQ-KLRRTHPDMALKIKSE 564
+L E+PD+F +++H IP P+ PP+ L L +K
Sbjct: 343 VLEEFPDLFNPEKAYQVPASRGLLDHAIPLIPDKDGNDPPLPWGPLYGMSRQELLVLKKT 522
Query: 565 V*KQIDAGFLMTIEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVD 624
+ +D GF+ A ++ V K G +R CVD+R LN + KD +PLP I +
Sbjct: 523 LEDLLDKGFIKASGSAAG-APVLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISETLR 699
Query: 625 NTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMT 684
A ++ F+ +D + ++++++ ED+EKT+F T +G F + V PFGL A AT+QR +
Sbjct: 700 RVAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQRYIN 879
Query: 685 TLFHDMIHKEVEVYVDDMIV-KSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGK 743
H+ + V Y+DD+++ + ++ H + ++ RL L L+P KC F V + K
Sbjct: 880 KTLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVTTVK 1059
Query: 744 LLGFIVSQ-KGIEVDPDKVRAIREMPAPQTEKQVRGFLRRLNYISRFISHMTATCGPIFK 802
+GFI++ KG+ DP K+ AIR+ P + K R FL NY FI + P+ +
Sbjct: 1060YVGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGARSFLGFCNYYKDFIPGYSEITEPLTR 1239
Query: 803 LLRKNQPIVWNDECQGAFDSIKNYLLEPPILVPPMEGKPLIMYLSVFDESVGCVLGQQDE 862
L RK+ P W E + AF +K E P+L + ++G VL Q+D
Sbjct: 1240LTRKDFPFRWGAEQEAAFTKLKRLFAEEPVLRMFDPEAVTTVETDCSGFALGGVLTQEDG 1419
Query: 863 TGKKEHAIYYLSKKFTDCETRYMMLEKTCCAPAWAAKR 900
TG H + + S++ + E Y + +K A WA R
Sbjct: 1420TG-AAHPVAFHSQRLSPAEYNYPIHDKELLA-VWACLR 1527
>AL375832
Length = 535
Score = 117 bits (292), Expect = 5e-26
Identities = 70/120 (58%), Positives = 76/120 (63%)
Frame = +2
Query: 289 VCQCNY*RGDWIWS*ACVCYTRSHCERLGCHRHSFNHACFRVMCLVA*VLFFQKNSNPLA 348
VC C *R +WIWS AC+CYTR HCE LG H HSFNHACFRVM LVA L FQK + +
Sbjct: 2 VC*CYL*RSNWIWSQACICYTRRHCE*LGRH*HSFNHACFRVMRLVALALCFQK*QSSRS 181
Query: 349 LPKAKVIFSVRA*LFRHSRYQ*KSSFLSRLLFIFFSAFSLEIMVMPEKTKTSVFSY*FQK 408
IF + +*K SFLSRLL FF L MVMPEKT TSVFSY*FQK
Sbjct: 182 AQSESDIFCKGVFVSALPFDE*KLSFLSRLL--FF*CLFLGKMVMPEKT*TSVFSY*FQK 355
Score = 66.6 bits (161), Expect = 7e-11
Identities = 53/134 (39%), Positives = 63/134 (46%), Gaps = 12/134 (8%)
Frame = +3
Query: 339 FFQKNSNPLALPKAKVIFSVRA*LFRHSRYQ*KSSFLSRLLFIFFSAFSLEIMVMPEKTK 398
F KN+NPLALPKA+VIFSVRA LFRH R K+ F FFSAF LE +K K
Sbjct: 150 FVFKNNNPLALPKARVIFSVRACLFRHCRLMNKNCRFFHDCF-FFSAFFLEKW*CQKKPK 326
Query: 399 TSVFSY*FQKICIKTSF*NHPTILRRLSHSKPVGHSNPTFP------------LNFEFPV 446
F F+ + K + + S+ H P FPV
Sbjct: 327 RLYFLINFKNLHEKKKIQKTKKLFLKSSN-----HYMQVEP**TR*TQ*FYASSKL*FPV 491
Query: 447 YEAEDEEGDDIPYE 460
YE + EGDDIPYE
Sbjct: 492 YEPKMREGDDIPYE 533
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 84.7 bits (208), Expect(2) = 4e-22
Identities = 37/45 (82%), Positives = 42/45 (93%)
Frame = -2
Query: 889 KTCCAPAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKPAVLLGRL 933
KTCCA AWAAKRLRHY++NHTTWL+S+MDPIKYIFEKPA L GR+
Sbjct: 635 KTCCALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPA-LTGRI 504
Score = 48.1 bits (113), Expect = 3e-05
Identities = 20/24 (83%), Positives = 23/24 (95%)
Frame = +2
Query: 855 CVLGQQDETGKKEHAIYYLSKKFT 878
CVLGQQDETG+KEHAIYYLS+K +
Sbjct: 38 CVLGQQDETGRKEHAIYYLSRKLS 109
Score = 40.0 bits (92), Expect(2) = 4e-22
Identities = 33/82 (40%), Positives = 37/82 (44%)
Frame = -3
Query: 933 LHAGRCSCLNMTLCSRLKNQSKVAFLPIILPTNLLMITNQLSSISPMKRSCISNQKTAKN 992
LH GRC NM + + Q K FL I TN L + S MKR CI K +
Sbjct: 505 LHGGRCYYRNMISSTVPRRQLKAVFLLITWLTNHLKTIDLSSLTFLMKRLCI*R*KIVTS 326
Query: 993 R*LVKVQIPIASGD*SLMVLSM 1014
KV I I G * LM LSM
Sbjct: 325 HYSEKVLIQIQCGV*YLMGLSM 260
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 85.5 bits (210), Expect = 1e-16
Identities = 66/230 (28%), Positives = 109/230 (46%), Gaps = 5/230 (2%)
Frame = +2
Query: 533 VEHRIPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKK 592
++ I P P+ R +P +K ++ ++ GF+ YP V ++ + KK
Sbjct: 35 IDFGIDLLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVV-VLFLKKK 211
Query: 593 DGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDRE 652
DG +RM +D+ LN + K +PLP ID L DN SK F +D G +Q ++ ED
Sbjct: 212 DGFLRMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVIGEDVP 391
Query: 653 KTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQH 712
KT+F +G + VM FG N + M +F D + V V+ +D+++ SK+E +H
Sbjct: 392 KTAFRIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSKNENEH 571
Query: 713 VEYLTKMFERLRKYKLRLNPNKCTFG-----VRSGKLLGFIVSQKGIEVD 757
+L + L+ L C V G ++S +G++VD
Sbjct: 572 ENHLRLALKVLKDIGL------CQISYV*ILVEVGFFSLHVISGEGLKVD 703
>BE942785
Length = 460
Score = 83.2 bits (204), Expect = 7e-16
Identities = 45/84 (53%), Positives = 52/84 (61%)
Frame = -1
Query: 185 VCKEWEACYSPWRRGIFG*PAVIFLLH*SWIC*RNFLSRFDY*RYGA*EGWGYYGFFKRR 244
+CK W ACY WR I *PA+IFLLH S +C + SR + RYGA EGW GFF+
Sbjct: 385 ICK-WXACYHSWRGSILD*PAIIFLLHRSRVCRGDRFSRINCRRYGAQEGWNCNGFFEGC 209
Query: 245 SEGCPRRIGCRLGQVNTTPRE*A* 268
+E PR CRLGQVNT E A*
Sbjct: 208 TESRPRESSCRLGQVNTAS*EQA* 137
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 48.5 bits (114), Expect = 2e-05
Identities = 22/49 (44%), Positives = 30/49 (60%)
Frame = -1
Query: 584 ANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVF 632
A ++ V KKDG RMC+D+R NK + K+ +PLP ID L D + F
Sbjct: 238 AALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCYF 92
>CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product {Drosophila
melanogaster}, partial (20%)
Length = 192
Score = 47.4 bits (111), Expect = 5e-05
Identities = 23/56 (41%), Positives = 30/56 (53%)
Frame = +1
Query: 850 DESVGCVLGQQDETGKKEHAIYYLSKKFTDCETRYMMLEKTCCAPAWAAKRLRHYL 905
D +G L Q+DE EH I Y S+ T E Y ++E+ C A WA + RHYL
Sbjct: 7 DWGIGAGLSQKDEENH-EHPIAYASRLLTAAERNYTVVERECLAAIWAIRNFRHYL 171
>BQ158004 weakly similar to SP|Q8VXP3|TBC2 Tbc2 translation factor
chloroplast precursor. {Chlamydomonas reinhardtii},
partial (1%)
Length = 958
Score = 32.3 bits (72), Expect = 1.5
Identities = 13/31 (41%), Positives = 15/31 (47%)
Frame = -3
Query: 304 ACVCYTRSHCERLGCHRHSFNHACFRVMCLV 334
AC CY S C L C R H C R C++
Sbjct: 680 ACYCY*SSLCVALSCRRLMVIHVCVRFFCIL 588
>BG448680 homologue to GP|4249411|gb|A unknown protein {Arabidopsis thaliana},
partial (29%)
Length = 659
Score = 32.0 bits (71), Expect = 2.0
Identities = 19/50 (38%), Positives = 27/50 (54%), Gaps = 5/50 (10%)
Frame = -3
Query: 1530 IEKEDKPTTRPKGQV---DA*L*GSLCCQEGLLWWCFNSY--THGWCRTS 1574
+E ++ P ++ KG V DA L + C + LWWCF Y H C+TS
Sbjct: 168 LESDEAPNSQNKGYVKPPDAEL--AECVTKNTLWWCFLRYYDCHRSCQTS 25
>BG584039 homologue to GP|4249411|gb|A unknown protein {Arabidopsis thaliana},
partial (23%)
Length = 800
Score = 32.0 bits (71), Expect = 2.0
Identities = 19/50 (38%), Positives = 27/50 (54%), Gaps = 5/50 (10%)
Frame = -1
Query: 1530 IEKEDKPTTRPKGQV---DA*L*GSLCCQEGLLWWCFNSY--THGWCRTS 1574
+E ++ P ++ KG V DA L + C + LWWCF Y H C+TS
Sbjct: 212 LESDEAPNSQNKGYVKPPDAEL--AECVTKNTLWWCFLRYYDCHRSCQTS 69
>TC77920 similar to GP|15809948|gb|AAL06901.1 AT3g10210/F14P13_19
{Arabidopsis thaliana}, partial (78%)
Length = 1156
Score = 31.6 bits (70), Expect = 2.6
Identities = 11/29 (37%), Positives = 17/29 (57%)
Frame = -2
Query: 827 LLEPPILVPPMEGKPLIMYLSVFDESVGC 855
+L PP+ +PPM P +++ SV GC
Sbjct: 117 MLRPPLRIPPMNSSPPLLFFSVIPVQRGC 31
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.346 0.153 0.549
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 55,902,271
Number of Sequences: 36976
Number of extensions: 933222
Number of successful extensions: 8661
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 3425
Number of HSP's successfully gapped in prelim test: 400
Number of HSP's that attempted gapping in prelim test: 4767
Number of HSP's gapped (non-prelim): 4559
length of query: 1586
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1477
effective length of database: 4,984,343
effective search space: 7361874611
effective search space used: 7361874611
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC147714.3


