

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147712.15 + phase: 0 /pseudo/partial
(740 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
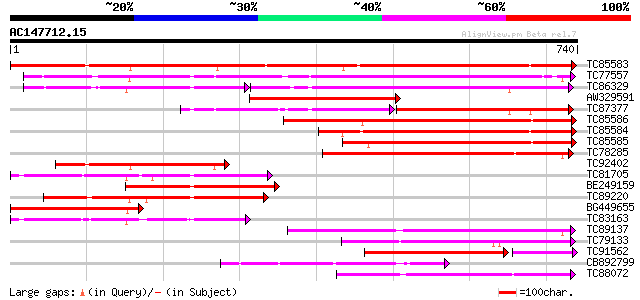
Score E
Sequences producing significant alignments: (bits) Value
TC85583 similar to PIR|T51335|T51335 subtilisin-like proteinase ... 755 0.0
TC77557 similar to PIR|JC7519|JC7519 subtilisin-like serine prot... 506 e-144
TC86329 similar to PIR|T07171|T07171 subtilisin-like proteinase ... 315 e-142
AW329591 weakly similar to GP|15010692|gb| At2g04160/T16B23.1 {A... 390 e-108
TC87377 weakly similar to PIR|T07171|T07171 subtilisin-like prot... 207 e-104
TC85586 similar to GP|20521377|dbj|BAB91889. putative subtilisin... 320 2e-87
TC85584 weakly similar to PIR|A84473|A84473 probable serine prot... 292 3e-79
TC85585 similar to GP|22773236|gb|AAN06842.1 Putatvie subtilisin... 284 1e-76
TC78285 similar to PIR|A84473|A84473 probable serine proteinase ... 267 1e-71
TC92402 similar to GP|20198252|gb|AAM15483.1 subtilisin-like ser... 263 2e-70
TC81705 weakly similar to GP|6721520|dbj|BAA89562.1 EST AU029428... 261 7e-70
BE249159 weakly similar to GP|20198252|gb subtilisin-like serine... 247 1e-65
TC89220 similar to PIR|T05768|T05768 subtilisin-like proteinase ... 243 3e-64
BG449655 similar to GP|20198252|gb subtilisin-like serine protea... 241 6e-64
TC83163 weakly similar to GP|6721520|dbj|BAA89562.1 EST AU029428... 235 4e-62
TC89137 similar to GP|4200338|emb|CAA76726.1 P69C protein {Lycop... 228 8e-60
TC79133 similar to GP|14150446|gb|AAK53065.1 subtilisin-type pro... 218 9e-57
TC91562 similar to PIR|T07172|T07172 subtilisin-like proteinase ... 179 4e-55
CB892799 weakly similar to PIR|T05768|T05 subtilisin-like protei... 211 8e-55
TC88072 similar to PIR|T06580|T06580 subtilisin-like proteinase ... 208 7e-54
>TC85583 similar to PIR|T51335|T51335 subtilisin-like proteinase AIR3
auxin-induced [imported] - Arabidopsis thaliana
(fragment), partial (40%)
Length = 2691
Score = 755 bits (1949), Expect = 0.0
Identities = 411/776 (52%), Positives = 522/776 (66%), Gaps = 38/776 (4%)
Frame = +2
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLE 61
YIVY+G+HSHGP P++ +L+ AT+SHY+LL S LGS EKAKEAI YSYNKHINGFAA+LE
Sbjct: 122 YIVYLGAHSHGPRPTSLELEIATNSHYDLLSSTLGSREKAKEAIIYSYNKHINGFAALLE 301
Query: 62 VEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTIIA 121
EEAA IAK NVVSVF +K H+L TTRSWEFLGL N K++ W+KG++GE TIIA
Sbjct: 302 DEEAADIAKKRNVVSVFLSKPHKLHTTRSWEFLGLRRN----AKNTAWQKGKFGENTIIA 469
Query: 122 NIDSGVSPESKSFSDDGMGPVPSRWRG--ICQLDNFH------CNRKLIGARFYSQGYES 173
NID+GV PESKSF+D G GPVPS+WRG C++ F CNRKLIGARF+S YE+
Sbjct: 470 NIDTGVWPESKSFNDKGYGPVPSKWRGGKACEISKFSKYKKNPCNRKLIGARFFSNAYEA 649
Query: 174 KFGRLNQSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCW 233
+L ARD LGHGT TLS AGGNFV A+VF + NGT KGGSPR+ VA YKVCW
Sbjct: 650 YNDKLPSWQRTARDFLGHGTHTLSTAGGNFVPDASVFAIGNGTVKGGSPRARVATYKVCW 829
Query: 234 LGTI*IECTDADIMQAFEDAISDGVDIISCSLGQTS---PKEFFEDGISIGAFHAIENGV 290
+C AD++ A + AISDGVDIIS SL S P++ F D +SIGAFHA+ +
Sbjct: 830 SLLDLEDCFGADVLAAIDQAISDGVDIISLSLAGHSLVYPEDIFTDEVSIGAFHALSRNI 1009
Query: 291 IVVAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPNEK 350
++VA GN GP G+V NVAPW+F++AAST+DR+F S + +G++ I G SL LP +
Sbjct: 1010LLVASAGNEGPTGGSVVNVAPWVFTIAASTLDRDFSSTITIGNQ-TIRGASLFVNLPPNQ 1186
Query: 351 FYSLVSSVDAKVGNATIEDAKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGS 410
+ L+ S D K+ NAT DA+ CK G+LDP+KVKGKI+ C+ V +EA+S G+
Sbjct: 1187AFPLIVSTDGKLANATNHDAQFCKPGTLDPSKVKGKIVECIREGNIKSVAEGQEALSAGA 1366
Query: 411 IGLVLGNDKQRGNDIMAYAHLLPT-----------------------SHINYTDGEYVHS 447
G++L N ++G +A H L SH D + S
Sbjct: 1367KGMLLSNQPKQGKTTLAEPHTLSCVEVPHHAPKPPKPKKSAEQERAGSHAPAFDITSMDS 1546
Query: 448 YIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIG 507
+KA T + AKT G KPAPV+AS SSRGPN IQP ILKPD+TAPGV+IL AY
Sbjct: 1547KLKAGTT--IKFSGAKTLYGRKPAPVMASFSSRGPNKIQPSILKPDVTAPGVNILAAYSL 1720
Query: 508 AISPTGLASDNQ-WIPYNIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQ 566
S + L +DN+ P+N+ GTS+SCPHV+ I L+KT++PNWSPAA KSAIMTT T
Sbjct: 1721YASASNLKTDNRNNFPFNVLQGTSMSCPHVAGIAGLIKTLHPNWSPAAIKSAIMTTATTL 1900
Query: 567 GNNHRPIKDQSKED-ATPFGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMK 625
N +RPI+D + A PF YG+GH+QP+LA+DPGLVYDL I DYLNFLCA+GYNQ +
Sbjct: 1901DNTNRPIQDAFENKLAIPFDYGSGHVQPDLAIDPGLVYDLGIKDYLNFLCAYGYNQQLIS 2080
Query: 626 MFS-RKPYICPKSYNMLDFNYPSITVPNLGKHFVQEVTRTVTNVGSPGTYRVQVNEPHGI 684
+ +IC S+++ DFNYPSIT+PNL + V VTRTVTNVG PGTY + + G
Sbjct: 2081ALNFNGTFICSGSHSITDFNYPSITLPNLKLNAV-NVTRTVTNVGPPGTYSAKA-QLLGY 2254
Query: 685 FVLIKPRSLTFNEVGEKKTFKIIFKVTKPTSSG-YVFGHLLWSDGRHKVMSPLVVK 739
+++ P SLTF + GEKKTF++I + T T G Y FG+L W+DG+H V SP+ V+
Sbjct: 2255KIVVLPNSLTFKKTGEKKTFQVIVQATNVTPRGKYQFGNLQWTDGKHIVRSPITVR 2422
>TC77557 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase (EC
3.4.21.-) - Arabidopsis thaliana, partial (88%)
Length = 2682
Score = 506 bits (1304), Expect = e-144
Identities = 293/742 (39%), Positives = 439/742 (58%), Gaps = 21/742 (2%)
Frame = +3
Query: 18 SDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLEVEEAAKIAKHPNVVSV 77
S++ + + H S L S + E + Y+Y I+GF+ L EEA + +++V
Sbjct: 351 SEMPESFEHHTLWYESSLQSVSDSAE-MMYTYENAIHGFSTRLTPEEARLLESQTGILAV 527
Query: 78 FENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTIIANIDSGVSPESKSFSDD 137
+EL TTR+ +FLGL+ + + P+ S G ++ +D+GV PESKSF+D
Sbjct: 528 LPEVKYELHTTRTPQFLGLDKSADMFPESSS------GNEVVVGVLDTGVWPESKSFNDA 689
Query: 138 GMGPVPSRWRGICQLD-NF---HCNRKLIGARFYSQGYESKFGRLNQSLYNA--RDVLGH 191
G GP+P+ W+G C+ NF +CN+KLIGARF+S+G E+ G ++++ + RD GH
Sbjct: 690 GFGPIPTTWKGACESGTNFTAANCNKKLIGARFFSKGVEAMLGPIDETTESKSPRDDDGH 869
Query: 192 GTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI*IECTDADIMQAFE 251
GT T S A G+ V A++FG A+GTA+G + R+ VA YKVCW G C +DI+ A +
Sbjct: 870 GTHTSSTAAGSVVPDASLFGYASGTARGMATRARVAVYKVCWKGG----CFSSDILAAID 1037
Query: 252 DAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGNSGPKFGTVTNVAP 311
AISD V+++S SLG ++F D ++IGAF A+E G++V GN+GP +++NVAP
Sbjct: 1038 KAISDNVNVLSLSLGG-GMSDYFRDSVAIGAFSAMEKGILVSCSAGNAGPSAYSLSNVAP 1214
Query: 312 WLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTG--LPNEKFYSLVSSVDAKVGNAT-IE 368
W+ +V A T+DR+F + + LG+ G SL G LP + + GNAT
Sbjct: 1215 WITTVGAGTLDRDFPASVSLGNGLNYSGVSLYRGNALPESPLPLIYA------GNATNAT 1376
Query: 369 DAKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVLGNDKQRGNDIMAY 428
+ +C G+L P V GKI+ C R ++ V + G +G+VL N G +++A
Sbjct: 1377 NGNLCMTGTLSPELVAGKIVLCD-RGMNARVQKGAVVKAAGGLGMVLSNTAANGEELVAD 1553
Query: 429 AHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPI 488
HLLP + + +G + Y+ + P + T+VGV+P+PV+A+ SSRGPN I P
Sbjct: 1554 THLLPATAVGEREGNAIKKYLFSEAKPTVKIVFQGTKVGVEPSPVVAAFSSRGPNSITPQ 1733
Query: 489 ILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYP 548
ILKPD+ APGV+IL + A+ PTGLA D + + +NI SGTS+SCPHVS + AL+K+ +P
Sbjct: 1734 ILKPDLIAPGVNILAGWSKAVGPTGLAVDERRVDFNIISGTSMSCPHVSGLAALIKSAHP 1913
Query: 549 NWSPAAFKSAIMTTTTIQGNNHRPIKDQSK-EDATPFGYGAGHIQPELAMDPGLVYDLNI 607
+WSPAA +SA+MTT I N ++D + + +TPF +G+GH+ P A++PGLVYDL
Sbjct: 1914 DWSPAAVRSALMTTAYIAYKNGNKLQDSATGKSSTPFDHGSGHVDPVAALNPGLVYDLTA 2093
Query: 608 VDYLNFLCAHGYNQTQMKMFSRKPYICP--KSYNMLDFNYPSITV--PNLGKHFVQEVTR 663
DYL FLCA Y TQ+ +R+ + C K Y++ D NYPS V +G V + TR
Sbjct: 2094 DDYLGFLCALNYTATQITSLARRKFQCDAGKKYSVSDLNYPSFAVVFDTMGGANVVKHTR 2273
Query: 664 TVTNVGSPGTYRVQV-NEPHGIFVLIKPRSLTFNEVGEKKTFKIIFKVTKPTSSGYV--- 719
TNVG GTY+ V ++ + + ++P L+F + EKK+F + F TSSG
Sbjct: 2274 IFTNVGPAGTYKASVTSDSKNVKITVEPEELSF-KANEKKSFTVTF-----TSSGSTPQK 2435
Query: 720 ---FGHLLWSDGRHKVMSPLVV 738
FG L W++G++ V SP+ +
Sbjct: 2436 LNGFGRLEWTNGKNVVGSPISI 2501
>TC86329 similar to PIR|T07171|T07171 subtilisin-like proteinase (EC 3.4.21.-)
1 - tomato, partial (91%)
Length = 2671
Score = 315 bits (807), Expect(2) = e-142
Identities = 176/437 (40%), Positives = 257/437 (58%), Gaps = 15/437 (3%)
Frame = +3
Query: 315 SVAASTIDRNFVSYLQLGDKHIIMGTSLSTG-LPNEKFYSLVSSVDAKVGNATIEDAKIC 373
+V A TIDR+F +Y+ LG+ + G SL G LP LV + + ++ +C
Sbjct: 1086 TVGARTIDRDFPAYITLGNGNRYNGVSLYNGKLPPNSPLPLVYAANVSQDSSD----NLC 1253
Query: 374 KVGSLDPNKVKGKILFCLLRELDGLVYAEEEAI--SGGSIGLVLGNDKQRGNDIMAYAHL 431
SL P+KV GKI+ C + G AE+ + G IG++L N++ G +++A + L
Sbjct: 1254 STDSLIPSKVSGKIVIC---DRGGNPRAEKSLVVKRAGGIGMILANNQDYGEELVADSFL 1424
Query: 432 LPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPIILK 491
LP + + + Y + P A + T GV+P+PV+A+ SSRGPN + P ILK
Sbjct: 1425 LPAAALGEKASNEIKKYASSAPNPTAKIAFGGTRFGVQPSPVVAAFSSRGPNILTPKILK 1604
Query: 492 PDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWS 551
PD+ APGV+IL + G + PTGL+ D + + +NI SGTS+SCPHVS + ALLK +P WS
Sbjct: 1605 PDLIAPGVNILAGWSGKVGPTGLSVDTRHVSFNIISGTSMSCPHVSGLAALLKGAHPEWS 1784
Query: 552 PAAFKSAIMTTTTIQGNNHRPIKDQSKE-DATPFGYGAGHIQPELAMDPGLVYDLNIVDY 610
PAA +SA+MTT+ N + IKD + ATP YG+GH+ P A+DPGLVYD DY
Sbjct: 1785 PAAIRSALMTTSYGTYKNGQTIKDVATGIPATPLDYGSGHVDPVAALDPGLVYDATTDDY 1964
Query: 611 LNFLCAHGYNQTQMKMFSRKPYICPK--SYNMLDFNYPSITVP--------NLGKHFVQE 660
LNFLCA YN Q+K+ +R+ + C K Y + D NYPS +VP + + +
Sbjct: 1965 LNFLCALNYNSFQIKLVARREFTCDKRIKYRVEDLNYPSFSVPFDTASGRGSSHNPSIVQ 2144
Query: 661 VTRTVTNVGSPGTYRVQVNEPHGI-FVLIKPRSLTFNEVGEKKTFKIIFKVTKPTSSGYV 719
R +TNVG+P TY+V V+ + ++++P++L+F E+ EKK++ + F S
Sbjct: 2145 YKRILTNVGAPSTYKVSVSSQSPLDKIVVEPQTLSFKELNEKKSYTVTFTSHSMPSGTTS 2324
Query: 720 FGHLLWSDGRHKVMSPL 736
F HL WSDG+HKV SP+
Sbjct: 2325 FAHLEWSDGKHKVTSPI 2375
Score = 208 bits (529), Expect(2) = e-142
Identities = 115/303 (37%), Positives = 181/303 (58%), Gaps = 8/303 (2%)
Frame = +2
Query: 19 DLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLEVEEAAKIAKHPNVVSVF 78
++ ++ D H S L S E + Y+Y +GF+ L +EA + K P ++SV
Sbjct: 206 NMPASFDDHLQWYDSSLKSVSDTAETM-YTYKHVAHGFSTRLTTQEADLLTKQPGILSVI 382
Query: 79 ENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTIIANIDSGVSPESKSFSDDG 138
+ +EL TTR+ EFLGLE ++P G+ E I+ ID+GV PE KSF D G
Sbjct: 383 PDVRYELHTTRTPEFLGLEKTITLLPSS-----GKQSE-VIVGVIDTGVWPELKSFDDTG 544
Query: 139 MGPVPSRWRGICQ----LDNFHCNRKLIGARFYSQGYESKFGRLNQSLYNA--RDVLGHG 192
+GPVP W+G C+ ++ +CN+KL+GARF+++GYE+ FG ++++ + RD GHG
Sbjct: 545 LGPVPKSWKGECETGKTFNSSNCNKKLVGARFFAKGYEAAFGPIDENTESKSPRDDDGHG 724
Query: 193 TPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI*IECTDADIMQAFED 252
+ T + A G+ V+GA++FG A+GTAKG + ++ VAAYKVCWLG C +DI A +
Sbjct: 725 SHTSTTAAGSAVAGASLFGFASGTAKGMATQARVAAYKVCWLG----GCFTSDIAAAIDK 892
Query: 253 AISDGVDIISCSLGQTSP--KEFFEDGISIGAFHAIENGVIVVAGGGNSGPKFGTVTNVA 310
AI + + + ++++D +++G F AIE+G++V + GN GP ++ NVA
Sbjct: 893 AIDTQARTAAYKVSRLGGGLTDYYKDTVAMGTFAAIEHGILVSSSAGNGGPSKASLANVA 1072
Query: 311 PWL 313
PW+
Sbjct: 1073PWI 1081
>AW329591 weakly similar to GP|15010692|gb| At2g04160/T16B23.1 {Arabidopsis
thaliana}, partial (22%)
Length = 592
Score = 390 bits (1002), Expect = e-108
Identities = 197/197 (100%), Positives = 197/197 (100%)
Frame = +2
Query: 314 FSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPNEKFYSLVSSVDAKVGNATIEDAKIC 373
FSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPNEKFYSLVSSVDAKVGNATIEDAKIC
Sbjct: 2 FSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPNEKFYSLVSSVDAKVGNATIEDAKIC 181
Query: 374 KVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVLGNDKQRGNDIMAYAHLLP 433
KVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVLGNDKQRGNDIMAYAHLLP
Sbjct: 182 KVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVLGNDKQRGNDIMAYAHLLP 361
Query: 434 TSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPIILKPD 493
TSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPIILKPD
Sbjct: 362 TSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPIILKPD 541
Query: 494 ITAPGVDILYAYIGAIS 510
ITAPGVDILYAYIGAIS
Sbjct: 542 ITAPGVDILYAYIGAIS 592
>TC87377 weakly similar to PIR|T07171|T07171 subtilisin-like proteinase (EC
3.4.21.-) 1 - tomato, partial (54%)
Length = 2071
Score = 207 bits (526), Expect(2) = e-104
Identities = 107/248 (43%), Positives = 150/248 (60%), Gaps = 16/248 (6%)
Frame = +1
Query: 505 YIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTT 564
+ GA+ PTGLA D + + +NI SGTS+SCPH S + A++K YP WSPAA +SA+MTT
Sbjct: 829 WTGAVGPTGLALDKRHVNFNIISGTSMSCPHASGLAAIVKGAYPEWSPAAIRSALMTTAY 1008
Query: 565 IQGNNHRPIKD-QSKEDATPFGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQ 623
N + I D + + ATPF +G+GH+ P A+DPGLVYD+N+ DYL F CA Y Q
Sbjct: 1009 TSYKNGQTIVDVATGKPATPFDFGSGHVDPVSALDPGLVYDINVDDYLGFFCALNYTSYQ 1188
Query: 624 MKMFSRKPYICP--KSYNMLDFNYPSITVP---------NLGKHFVQEVTRTVTNVGSPG 672
+K+ +R+ + C K Y + DFNYPS V K + E R +TNVG+PG
Sbjct: 1189 IKLAARREFTCDARKKYRVEDFNYPSFAVALETASGIGGGSNKPIIVEYNRVLTNVGAPG 1368
Query: 673 TYRVQV----NEPHGIFVLIKPRSLTFNEVGEKKTFKIIFKVTKPTSSGYVFGHLLWSDG 728
TY V + + V+++P +++F EV EKK +K+ F S FG+L W+DG
Sbjct: 1369 TYNATVVLSSVDSSSVKVVVEPETISFKEVYEKKGYKVRFICGSMPSGTKSFGYLEWNDG 1548
Query: 729 RHKVMSPL 736
+HKV SP+
Sbjct: 1549 KHKVGSPI 1572
Score = 190 bits (483), Expect(2) = e-104
Identities = 113/282 (40%), Positives = 167/282 (59%), Gaps = 3/282 (1%)
Frame = +2
Query: 224 SHVAAYKVCWLGTI*IECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAF 283
+ VAAYKVCWL C +DI + AI DGV+I+S S+G S +++ D I+IGAF
Sbjct: 11 ARVAAYKVCWLSG----CFTSDIAAGMDKAIEDGVNILSMSIGG-SIMDYYRDIIAIGAF 175
Query: 284 HAIENGVIVVAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLS 343
A+ +G++V + GN GP +++NVAPW+ +V A TIDR+F SY+ LG+ G SL
Sbjct: 176 TAMSHGILVSSSAGNGGPSAESLSNVAPWITTVGAGTIDRDFPSYITLGNGKTYTGASLY 355
Query: 344 TGLPNEKFYSLVSSVDAKVGNATIEDAK-ICKVGSLDPNKVKGKILFCLLRELDGLVYAE 402
G P+ SL+ V A GN + +C SL +KV GKI+ C E G E
Sbjct: 356 NGKPSSD--SLLPVVYA--GNVSESSVGYLCIPDSLTSSKVLGKIVIC---ERGGNSRVE 514
Query: 403 EEAI--SGGSIGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMT 460
+ + + G +G++L N++ G +++A +HLLP + + + Y+ TK P A +
Sbjct: 515 KGLVVKNAGGVGMILVNNEAYGEELIADSHLLPAAALGQKSSTVLKDYVFTTKNPRAKLV 694
Query: 461 KAKTEVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDIL 502
T + V+P+PV+A+ SSRGPN + P ILKPD+ APGV+IL
Sbjct: 695 FGGTHLQVQPSPVVAAFSSRGPNSLTPKILKPDLIAPGVNIL 820
>TC85586 similar to GP|20521377|dbj|BAB91889. putative subtilisin-like
protease {Oryza sativa (japonica cultivar-group)},
partial (17%)
Length = 1460
Score = 320 bits (819), Expect = 2e-87
Identities = 183/394 (46%), Positives = 245/394 (61%), Gaps = 12/394 (3%)
Frame = +1
Query: 358 VDAKVGNATIEDAKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVLGN 417
+DAK NAT DA+ C+ +LDP+KVKGKI+ C V +EA+S G+ G+ L N
Sbjct: 1 IDAKFSNATTRDARFCRPRTLDPSKVKGKIVACAREGKIKSVAEGQEALSAGAKGMFLEN 180
Query: 418 D-KQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAY------MTKAKTEVGVKP 470
K GN +++ H+L T N + T T ++A T +G KP
Sbjct: 181 QPKVSGNTLLSEPHVLSTVGGNGQAAITAPPRLGVTATDTIESGTKIRFSQAITLIGRKP 360
Query: 471 APVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQ-WIPYNIGSGT 529
APV+AS SSRGPN +QP ILKPD+TAPGV+IL AY S + L +DN+ P+N+ GT
Sbjct: 361 APVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGT 540
Query: 530 SISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQ-SKEDATPFGYGA 588
S+SCPHV+ L+KT++PNWSPAA KSAIMTT T + N ++PI D K A PF YG+
Sbjct: 541 SMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATTRDNTNKPISDAFDKTLADPFAYGS 720
Query: 589 G-HIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFS-RKPYICPKSYNMLDFNYP 646
G + P A+DPGLVYDL I DYLNFLCA GYN+ + + + C ++++ D NYP
Sbjct: 721 GTYSHPTSAIDPGLVYDLGIKDYLNFLCASGYNKQLISALNFNMTFTCSGTHSIDDLNYP 900
Query: 647 SITVPNLGKHFVQEVTRTVTNVGSPGTYRVQVNEPHGIFVLIKPRSLTFNEVGEKKTFKI 706
SIT+PNLG + + VTRTVTNVG P TY +V P G + + P SL F ++GEKKTF++
Sbjct: 901 SITLPNLGLNAI-TVTRTVTNVGPPSTYFAKVQLP-GYKIAVVPSSLNFKKIGEKKTFQV 1074
Query: 707 IFKVTKP-TSSGYVFGHLLWSDGRHKVMSPLVVK 739
I + T Y FG L W++G+H V SP+ V+
Sbjct: 1075IVQATSEIPRRKYQFGELRWTNGKHIVRSPVTVQ 1176
>TC85584 weakly similar to PIR|A84473|A84473 probable serine proteinase
[imported] - Arabidopsis thaliana, partial (19%)
Length = 1241
Score = 292 bits (748), Expect = 3e-79
Identities = 170/350 (48%), Positives = 226/350 (64%), Gaps = 13/350 (3%)
Frame = +1
Query: 403 EEAISGGSIGLVLGNDKQ-RGNDIMAYAHLL-----PTSHINYTDG--EYVHSYIKA-TK 453
+EA+S G+ G++L N + G +++ H+L P +H T + + S IK+ TK
Sbjct: 25 QEALSAGAKGVILRNQPEINGKTLLSEPHVLSTISYPGNHSRTTGRSLDIIPSDIKSGTK 204
Query: 454 TPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTG 513
M+ AKT KPAPV+AS SSRGPN +QP ILKPD+TAPGV+IL AY S +
Sbjct: 205 LRMS---PAKTLNRRKPAPVMASYSSRGPNKVQPSILKPDVTAPGVNILAAYSLFASASN 375
Query: 514 LASDNQ-WIPYNIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRP 572
L +D + P+N+ GTS+SCPHV+ L+KT++PNWSPAA KSAIMTT T + N ++P
Sbjct: 376 LITDTRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATTRDNTNKP 555
Query: 573 IKDQ-SKEDATPFGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFS-RK 630
I D K A PF YG+GHI+P AMDPGLVYDL I DYLNFLCA GYNQ + +
Sbjct: 556 ISDAFDKTLANPFAYGSGHIRPNSAMDPGLVYDLGIKDYLNFLCASGYNQQLISALNFNM 735
Query: 631 PYICPKSYNMLDFNYPSITVPNLGKHFVQEVTRTVTNVGSPGTYRVQVNEPHGIFVLIKP 690
+ C + ++ D NYPSIT+PNLG + V VTRTVTNVG P TY +V + G + + P
Sbjct: 736 TFTCSGTSSIDDLNYPSITLPNLGLNSV-TVTRTVTNVGPPSTYFAKV-QLAGYKIAVVP 909
Query: 691 RSLTFNEVGEKKTFKIIFKVTKPT-SSGYVFGHLLWSDGRHKVMSPLVVK 739
SL F ++GEKKTF++I + T T Y FG L W++G+H V SP+ V+
Sbjct: 910 SSLNFKKIGEKKTFQVIVQATSVTPRRKYQFGELRWTNGKHIVRSPVTVR 1059
>TC85585 similar to GP|22773236|gb|AAN06842.1 Putatvie subtilisin-like
serine protease {Oryza sativa (japonica
cultivar-group)}, partial (21%)
Length = 1353
Score = 284 bits (726), Expect = 1e-76
Identities = 158/316 (50%), Positives = 200/316 (63%), Gaps = 11/316 (3%)
Frame = +2
Query: 435 SHINYTDGEYVHSYIKATKTPMAYMTKAKTEV-------GVKPAPVIASLSSRGPNPIQP 487
S INY D + + + TP T A + G KPAPV+AS SSRGPN +QP
Sbjct: 11 STINYHDPRSITTPKGSEITPEDIKTNATIRMSPANALNGRKPAPVMASFSSRGPNKVQP 190
Query: 488 IILKPDITAPGVDILYAYIGAISPTGLASDNQ-WIPYNIGSGTSISCPHVSAIVALLKTI 546
ILKPD+TAPGV+IL AY S + L +DN+ P+NI GTS+SCPHV L+KT+
Sbjct: 191 YILKPDVTAPGVNILAAYSLLASVSNLVTDNRRGFPFNIQQGTSMSCPHVVGTAGLIKTL 370
Query: 547 YPNWSPAAFKSAIMTTTTIQGNNHRPIKDQ-SKEDATPFGYGAGHIQPELAMDPGLVYDL 605
+PNWSPAA KSAIMTT T + N + PI+D A F YG+GHIQP A+DPGLVYDL
Sbjct: 371 HPNWSPAAIKSAIMTTATTRDNTNEPIEDAFENTTANAFAYGSGHIQPNSAIDPGLVYDL 550
Query: 606 NIVDYLNFLCAHGYNQTQM-KMFSRKPYICPKSYNMLDFNYPSITVPNLGKHFVQEVTRT 664
I DYLNFLCA GYNQ + + + C + ++ D NYPSIT+PNLG + V VTRT
Sbjct: 551 GIKDYLNFLCAAGYNQKLISSLIFNMTFTCYGTQSINDLNYPSITLPNLGLNAV-TVTRT 727
Query: 665 VTNVGSPGTYRVQVNEPHGIFVLIKPRSLTFNEVGEKKTFKIIFKVTKPTSSG-YVFGHL 723
VTNVG TY + P G +++ P SL F ++GEKKTFK+ + T T G Y FG L
Sbjct: 728 VTNVGPRSTYTAKAQLP-GYKIVVVPSSLKFKKIGEKKTFKVTVQATSVTPQGKYEFGEL 904
Query: 724 LWSDGRHKVMSPLVVK 739
WS+G+H V SP+ ++
Sbjct: 905 QWSNGKHIVRSPITLR 952
>TC78285 similar to PIR|A84473|A84473 probable serine proteinase [imported]
- Arabidopsis thaliana, partial (41%)
Length = 1353
Score = 267 bits (682), Expect = 1e-71
Identities = 137/335 (40%), Positives = 204/335 (60%), Gaps = 7/335 (2%)
Frame = +1
Query: 409 GSIGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGV 468
G IG++L N G +++A +HLLP + G+ + Y+ + P ++ T + V
Sbjct: 37 GGIGMILANTAASGEELVADSHLLPAVAVGRIIGDQIRKYVSSDLNPTTVLSFGGTVLNV 216
Query: 469 KPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSG 528
+P+PV+A+ SSRGPN I ILKPD+ PGV+IL + A+ P+GLA D + +NI SG
Sbjct: 217 RPSPVVAAFSSRGPNMITKEILKPDVIGPGVNILAGWSEAVGPSGLAEDTRKTKFNIMSG 396
Query: 529 TSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKED-ATPFGYG 587
TS+SCPH+S + ALLK +P WSP+A KSA+MTT N+ P++D + +TP +G
Sbjct: 397 TSMSCPHISGLAALLKAAHPTWSPSAIKSALMTTAYNHDNSKSPLRDAADGSFSTPLAHG 576
Query: 588 AGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYI-CPKSY-NMLDFNY 645
AGH+ P+ A+ PGLVYD + DY+ FLC+ YN Q+++ ++P + C K + N NY
Sbjct: 577 AGHVNPQKALSPGLVYDASTKDYITFLCSLNYNSEQIQLIVKRPSVNCTKKFANPGQLNY 756
Query: 646 PSITVPNLGKHFVQEVTRTVTNVGSPGT-YRVQVNEPHGIFVLIKPRSLTFNEVGEKKTF 704
PS +V K V+ TR VTNVG G+ Y V V+ P + + +KP L F +VGE+K +
Sbjct: 757 PSFSVVFSSKRVVR-YTRIVTNVGEAGSVYNVVVDVPSSVGITVKPSRLVFEKVGERKRY 933
Query: 705 KIIFKVTKPTSSGYV---FGHLLWSDGRHKVMSPL 736
+ F K + V FG +LWS+ +H+V SP+
Sbjct: 934 TVTFVSKKGADASKVRSGFGSILWSNAQHQVRSPI 1038
>TC92402 similar to GP|20198252|gb|AAM15483.1 subtilisin-like serine
protease AIR3 {Arabidopsis thaliana}, partial (16%)
Length = 704
Score = 263 bits (671), Expect = 2e-70
Identities = 136/237 (57%), Positives = 167/237 (70%), Gaps = 11/237 (4%)
Frame = +1
Query: 61 EVEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTII 120
E EEAA++AK+P VVSVF +K H+L TTRSWEFLGL N +S W+KGR+GE TII
Sbjct: 1 EEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGN----DINSAWQKGRFGENTII 168
Query: 121 ANIDSGVSPESKSFSDDGMGPVPSRWRG--ICQLDNFH------CNRKLIGARFYSQGYE 172
ANID+GV PES+SFSD G+GP+P++WRG +CQ++ CNRKLIGARF+S YE
Sbjct: 169 ANIDTGVWPESRSFSDRGIGPIPAKWRGGNVCQINKLRGSKKVPCNRKLIGARFFSDAYE 348
Query: 173 SKFGRLNQSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVC 232
G+L S ARD +GHGT TLS AGGNFV GA++F + NGT KGGSPR+ VA YKVC
Sbjct: 349 RYNGKLPTSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVC 528
Query: 233 WLGTI*IECTDADIMQAFEDAISDGVDIISCSLG---QTSPKEFFEDGISIGAFHAI 286
W T C AD++ A + AI DGVDII S G T+ +E F D +SIGAFHA+
Sbjct: 529 WSLTDAASCFGADVLSAIDQAIDDGVDIIFVSAGGPSSTNSEEIFTDEVSIGAFHAL 699
>TC81705 weakly similar to GP|6721520|dbj|BAA89562.1 EST AU029428(E30359)
corresponds to a region of the predicted gene.~Similar
to antifreeze-like, partial (33%)
Length = 1443
Score = 261 bits (667), Expect = 7e-70
Identities = 150/352 (42%), Positives = 208/352 (58%), Gaps = 10/352 (2%)
Frame = +1
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLE 61
Y+VY+G++ +G D Q H LL S + S E + + + YN +GF+A+L
Sbjct: 85 YVVYMGNNING-----EDDQIPESVHIELLSSIIPSEESERIKLIHHYNHAFSGFSAMLT 249
Query: 62 VEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTIIA 121
EA+ ++ H VVSVF + EL TTRSW+FL +++ G+ P ++ IIA
Sbjct: 250 QSEASALSGHDGVVSVFPDPILELHTTRSWDFL--DSDLGMKPSTNVLTHQHSSNDIIIA 423
Query: 122 NIDSGVSPESKSFSDDGMGPVPSRWRGICQ----LDNFHCNRKLIGARFYSQGYESKFGR 177
ID+G+ PES SF+D+G+G +PS W+GIC +CNRKLIGAR+Y+ + FG
Sbjct: 424 LIDTGIWPESPSFTDEGIGKIPSVWKGICMEGHDFKKSNCNRKLIGARYYNT--QDTFGS 597
Query: 178 LNQSLYNA----RDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCW 233
+ A RD +GHGT T S A G V+ AN +GLA GTA+GGSP + +AAYK C
Sbjct: 598 NKTHIGGAKGSPRDTVGHGTHTASTAAGVNVNNANYYGLAKGTARGGSPSTRIAAYKTCS 777
Query: 234 LGTI*IECTDADIMQAFEDAISDGVDIISCSLGQTS--PKEFFEDGISIGAFHAIENGVI 291
C+ + I++A +DAI DGVDIIS S+G +S ++ D I+IGAFHA + GV
Sbjct: 778 EEG----CSGSTILKAMDDAIKDGVDIISISIGLSSLMQSDYLNDPIAIGAFHAEQRGVT 945
Query: 292 VVAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLS 343
V GN GP TV N APW+F+VAAS IDRNF S + LG+ G ++
Sbjct: 946 AVCSAGNDGPDPNTVVNTAPWIFTVAASNIDRNFQSTIVLGNGKSFQGAGIN 1101
>BE249159 weakly similar to GP|20198252|gb subtilisin-like serine protease
AIR3 {Arabidopsis thaliana}, partial (24%)
Length = 661
Score = 247 bits (631), Expect = 1e-65
Identities = 124/202 (61%), Positives = 151/202 (74%), Gaps = 1/202 (0%)
Frame = +1
Query: 152 LDNFHCNRKLIGARFYSQGYESKFGRLNQSLYNARDVLGHGTPTLSVAGGNFVSGANVFG 211
L N +RKLIGAR + +GYE+ G+L+ S Y ARD +GHG+ TLS AGGNFV G +V+G
Sbjct: 64 LSNCSTSRKLIGARAFYKGYEAYVGKLDASFYTARDTIGHGSHTLSTAGGNFVQGVSVYG 243
Query: 212 LANGTAKGGSPRSHVAAYKVCWLGTI*IECTDADIMQAFEDAISDGVDIISCSLGQTSPK 271
NGTAKGGSP++HVAAYKVCW G C+DAD++ FE AISDGVD++S SLG +
Sbjct: 244 NGNGTAKGGSPKAHVAAYKVCWKG----GCSDADVLAGFEAAISDGVDVLSVSLGMKT-H 408
Query: 272 EFFEDGISIGAFHAIENGVIVVAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQL 331
F D ISIG+FHA+ NG++VVA GNSGP FGTV+NVAPWLF+VAASTIDR+F SY+ L
Sbjct: 409 NLFTDSISIGSFHAVANGIVVVASAGNSGPYFGTVSNVAPWLFTVAASTIDRDFASYVTL 588
Query: 332 GDKHIIMGTSLST-GLPNEKFY 352
GD GTSLS+ LP KFY
Sbjct: 589 GDNKHFKGTSLSSKDLPTHKFY 654
>TC89220 similar to PIR|T05768|T05768 subtilisin-like proteinase (EC
3.4.21.-) - Arabidopsis thaliana, partial (39%)
Length = 1149
Score = 243 bits (619), Expect = 3e-64
Identities = 128/308 (41%), Positives = 195/308 (62%), Gaps = 14/308 (4%)
Frame = +3
Query: 45 IFYSYNKHINGFAAVLEVEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVP 104
I ++Y+ +GF+AVL ++ A I+ HP++++VFE++ +L TTRS +FLGL N G
Sbjct: 228 ILHTYDTAFHGFSAVLTRQQVASISNHPSILAVFEDRRRQLHTTRSPQFLGLRNQRG--- 398
Query: 105 KDSIWEKGRYGEGTIIANIDSGVSPESKSFSDDGMGPVPSRWRGICQL-DNF---HCNRK 160
+W + YG I+ D+G+ PE +SFSD +GP+P RW+G+C+ + F +CNRK
Sbjct: 399 ---LWSESDYGSDVIVGVFDTGIWPERRSFSDMNLGPIPRRWKGVCESGEKFSPRNCNRK 569
Query: 161 LIGARFYSQGYESKFGR------LNQSLY--NARDVLGHGTPTLSVAGGNFVSGANVFGL 212
LIGAR++S+G+E G +N+++ + RD GHGT T S A G + AN+ G
Sbjct: 570 LIGARYFSKGHEVGAGSAGPLNPINETVEFRSPRDADGHGTHTASTAAGRYAFQANMSGY 749
Query: 213 ANGTAKGGSPRSHVAAYKVCWLGTI*IECTDADIMQAFEDAISDGVDIISCSLGQTS--P 270
A+G AKG +P++ +A YKVCW + C D+DI+ AF+ A++DGVD+IS S+G
Sbjct: 750 ASGIAKGVAPKARLAVYKVCWKNS---GCFDSDILAAFDAAVNDGVDVISISIGGGDGIA 920
Query: 271 KEFFEDGISIGAFHAIENGVIVVAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQ 330
++ D I+IG++ A+ GV V + GN GP +VTN+APWL +V A TIDR+F S +
Sbjct: 921 SPYYLDPIAIGSYGAVSRGVFVSSSAGNDGPSGMSVTNLAPWLTTVGAGTIDRDFPSQII 1100
Query: 331 LGDKHIIM 338
+ D I+
Sbjct: 1101IXDXKKIL 1124
>BG449655 similar to GP|20198252|gb subtilisin-like serine protease AIR3
{Arabidopsis thaliana}, partial (21%)
Length = 664
Score = 241 bits (616), Expect = 6e-64
Identities = 113/178 (63%), Positives = 139/178 (77%), Gaps = 4/178 (2%)
Frame = +1
Query: 1 SYIVYIGSHSHGPNPSAS-DLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAV 59
SY+VY+GSHSH +S D TDSHY LGS LGS + AKE+IFYSY +HINGFAA
Sbjct: 106 SYVVYLGSHSHDSEELSSVDFNRVTDSHYEFLGSFLGSSKTAKESIFYSYTRHINGFAAT 285
Query: 60 LEVEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTI 119
LE E AA+IAKHP V+SVFEN G +L TT SW F+GLE++YGV+P SIW K R+G+G I
Sbjct: 286 LEEEVAAEIAKHPKVLSVFENNGRKLHTTHSWGFMGLEDSYGVIPSSSIWNKARFGDGII 465
Query: 120 IANIDSGVSPESKSFSDDGMGPVPSRWRGICQL---DNFHCNRKLIGARFYSQGYESK 174
IAN+D+GV PESKSFSD+G GP+PS+WRGIC +FHCNRKLIGAR++++GY S+
Sbjct: 466 IANLDTGVWPESKSFSDEGFGPIPSKWRGICDKGRDPSFHCNRKLIGARYFNKGYASR 639
>TC83163 weakly similar to GP|6721520|dbj|BAA89562.1 EST AU029428(E30359)
corresponds to a region of the predicted gene.~Similar
to antifreeze-like, partial (29%)
Length = 990
Score = 235 bits (600), Expect = 4e-62
Identities = 138/319 (43%), Positives = 189/319 (58%), Gaps = 6/319 (1%)
Frame = +2
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLE 61
Y+VY+GS+ +G D Q H +LL S + S E + A+ + Y+ NGF+A+L
Sbjct: 95 YVVYMGSNINGV-----DGQIPESVHLDLLSSIIPSEESERIALIHHYSHAFNGFSAMLT 259
Query: 62 VEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTIIA 121
EA+ +A + VVSVFE+ EL TTRSW+F LE++ G+ P I + II
Sbjct: 260 QSEASALAGNDGVVSVFEDPFLELHTTRSWDF--LESDLGMRP-HGILKHQHSSNDIIIG 430
Query: 122 NIDSGVSPESKSFSDDGMGPVPSRWRGIC----QLDNFHCNRKLIGARFYSQGYESKFGR 177
ID+G+ PES SF D+G+G +PSRW+G+C +CNRKLIGAR+Y++
Sbjct: 431 VIDTGIWPESPSFKDEGIGKIPSRWKGVCMEAHDFKKSNCNRKLIGARYYNK-------- 586
Query: 178 LNQSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI 237
+ RD GHGT T S A G V+ A+ +GLA GTA+GGSP + +AAYK C G
Sbjct: 587 -KDPKGSPRDFNGHGTHTASTAAGVIVNNASYYGLAKGTARGGSPSARIAAYKAC-SGE- 757
Query: 238 *IECTDADIMQAFEDAISDGVDIISCSLGQTSP--KEFFEDGISIGAFHAIENGVIVVAG 295
C+ +++A +DAI DGVDIIS S+G +S E+ D I+IGAFHA + GV+VV
Sbjct: 758 --GCSGGTLLKAIDDAIKDGVDIISISIGFSSEFLSEYLSDPIAIGAFHAEQRGVMVVCS 931
Query: 296 GGNSGPKFGTVTNVAPWLF 314
GN GP TV N PW+F
Sbjct: 932 AGNEGPDHYTVVNTTPWIF 988
>TC89137 similar to GP|4200338|emb|CAA76726.1 P69C protein {Lycopersicon
esculentum}, partial (37%)
Length = 1230
Score = 228 bits (580), Expect = 8e-60
Identities = 143/384 (37%), Positives = 203/384 (52%), Gaps = 8/384 (2%)
Frame = +3
Query: 363 GNATIEDAKICKVGSLDPNKV--KGKILFCLLRELDGLVYAEEEAISGGSIGLVLGNDKQ 420
G+ D I G + KV KGKI+ C G V + G ++L N +
Sbjct: 12 GSINTSDDSIAFCGPIAMKKVDVKGKIVVCEQGGFVGRVAKGQAVKDAGGAAMILLNSEG 191
Query: 421 RGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSR 480
+ +A H+LP H++Y+ G + YI +T TPMA + T +G AP +AS SSR
Sbjct: 192 EDFNPIADVHVLPAVHVSYSAGLNIQDYINSTSTPMATILFKGTVIGNPNAPQVASFSSR 371
Query: 481 GPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIV 540
GP+ P ILKPDI PG++IL + ++ DN +NI +GTS+SCPH+S I
Sbjct: 372 GPSKASPGILKPDILGPGLNILAGW-------PISLDNSTSSFNIIAGTSMSCPHLSGIA 530
Query: 541 ALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKEDATPFGYGAGHIQPELAMDPG 600
ALLK +P+WSPAA KSAIMTT + +PI DQ A F GAGH+ P A DPG
Sbjct: 531 ALLKNSHPDWSPAAIKSAIMTTANHVNLHGKPILDQRLLPADVFATGAGHVNPSKANDPG 710
Query: 601 LVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYICP--KSYNMLDFNYPSITVPNLGKHFV 658
LVYD+ DY+ +LC Y Q+ + ++ C KS NYPSI++ LG +
Sbjct: 711 LVYDIETNDYVPYLCGLNYTDIQVGIILQQKVKCSDVKSIPQAQLNYPSISI-RLG-NTS 884
Query: 659 QEVTRTVTNVGSPG-TYRVQVNEPHGIFVLIKPRSLTFNEVGEKKTFKIIFKVTKPTSSG 717
Q +RT+TNVG TY V ++ P + + ++P +TF EV +K T+ + F + G
Sbjct: 885 QFYSRTLTNVGPVNTTYNVVIDVPVAVRMSVRPSQITFTEVKQKVTYWVDFIPEDKENRG 1064
Query: 718 YVF---GHLLWSDGRHKVMSPLVV 738
F G + W ++ V P+ V
Sbjct: 1065DNFIAQGSIKWISAKYSVSIPIAV 1136
>TC79133 similar to GP|14150446|gb|AAK53065.1 subtilisin-type protease
precursor {Glycine max}, partial (42%)
Length = 1101
Score = 218 bits (554), Expect = 9e-57
Identities = 121/314 (38%), Positives = 176/314 (55%), Gaps = 8/314 (2%)
Frame = +1
Query: 433 PTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPIILKP 492
P + + D + Y+ +T P+A + T + KPAP++A SSRGP+ + ILKP
Sbjct: 37 PATVVRSKDVVTLLKYVNSTSNPVATILPTVTVIDYKPAPMVAIFSSRGPSALSKNILKP 216
Query: 493 DITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSP 552
DI APGV IL A+IG + + +PY + +GTS+SCPHVS + +K+ P WS
Sbjct: 217 DIAAPGVTILAAWIGN-DDENVPKGKKPLPYKLETGTSMSCPHVSGLAGSIKSRNPTWSA 393
Query: 553 AAFKSAIMTTTTIQGNNHRPIKDQSKEDATPFGYGAGHIQPELAMDPGLVYDLNIVDYLN 612
+A +SAIMT+ T N PI ATP+ YGAG I + PGLVY+ + +DYLN
Sbjct: 394 SAIRSAIMTSATQINNMKAPITTDLGSVATPYDYGAGDITTIESFQPGLVYETSTIDYLN 573
Query: 613 FLCAHGYNQTQMKMFSR---KPYICPKSY---NMLDFNYPSITVPNLGKHFVQEVTRTVT 666
+LC GYN T +K+ S+ + CPK ++ + NYPSI + N V+RTVT
Sbjct: 574 YLCYIGYNTTTIKVISKTVPDTFNCPKESTPDHISNINYPSIAISNFTGKETVNVSRTVT 753
Query: 667 NVGSPG--TYRVQVNEPHGIFVLIKPRSLTFNEVGEKKTFKIIFKVTKPTSSGYVFGHLL 724
NVG Y VN P G+ V + P L F + +K++++ IF T + +FG +
Sbjct: 754 NVGEEDEVAYSAIVNAPSGVKVQLIPEKLQFTKSNKKQSYQAIFSTTLTSLKEDLFGSIT 933
Query: 725 WSDGRHKVMSPLVV 738
WS+G++ V SP V+
Sbjct: 934 WSNGKYSVRSPFVL 975
>TC91562 similar to PIR|T07172|T07172 subtilisin-like proteinase (EC
3.4.21.-) 2 - tomato, partial (36%)
Length = 1062
Score = 179 bits (454), Expect(2) = 4e-55
Identities = 89/190 (46%), Positives = 128/190 (66%), Gaps = 3/190 (1%)
Frame = +2
Query: 464 TEVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPY 523
T +G++P+P++A+ SSRGP+ + ILKPDI APGV+IL A+ G P+ L D++ + +
Sbjct: 29 TRLGIRPSPIVAAFSSRGPSLLTLEILKPDIVAPGVNILAAWSGLTGPSSLPIDHRRVKF 208
Query: 524 NIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKD-QSKEDAT 582
NI SGTS+SCPHVS I A++K +P WSPAA KSAIMTT + N +P++D S E +T
Sbjct: 209 NILSGTSMSCPHVSGIAAMIKAKHPEWSPAAIKSAIMTTAYVHDNTIKPLRDASSAEFST 388
Query: 583 PFGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYICPKS--YNM 640
P+ +GAGHI P A+DPGL+YD+ DY FLC + +++ +FS+ K +
Sbjct: 389 PYDHGAGHINPRKALDPGLLYDIEPQDYFEFLCTKKLSPSELVVFSKNSNRNCKHTLASA 568
Query: 641 LDFNYPSITV 650
D NYP+I+V
Sbjct: 569 SDLNYPAISV 598
Score = 54.7 bits (130), Expect(2) = 4e-55
Identities = 34/85 (40%), Positives = 46/85 (54%), Gaps = 1/85 (1%)
Frame = +1
Query: 657 FVQEVTRTVTNVG-SPGTYRVQVNEPHGIFVLIKPRSLTFNEVGEKKTFKIIFKVTKPTS 715
F + RTVTNVG + Y V G V ++P +L F +K ++KI FKVT S
Sbjct: 622 FCFNIHRTVTNVGPAVSKYHVIGTPFKGAVVKVEPDTLNFTRKYQKLSYKISFKVTSRQS 801
Query: 716 SGYVFGHLLWSDGRHKVMSPLVVKH 740
FG L+W D HKV SP+V+ +
Sbjct: 802 EPE-FGGLVWKDRLHKVRSPIVITY 873
>CB892799 weakly similar to PIR|T05768|T05 subtilisin-like proteinase (EC
3.4.21.-) - Arabidopsis thaliana, partial (20%)
Length = 875
Score = 211 bits (537), Expect = 8e-55
Identities = 122/300 (40%), Positives = 181/300 (59%), Gaps = 2/300 (0%)
Frame = +2
Query: 276 DGISIGAFHAIENGVIVVAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKH 335
D ISIG+FHA G+ VV+ GN G G+ TN+APW+ +VAA + DR+F S + LG+
Sbjct: 2 DAISIGSFHAANRGLFVVSSAGNEG-NLGSATNLAPWMLTVAAGSTDRDFTSDIILGNGA 178
Query: 336 IIMGTSLSTGLPNEKFYSLVSSVDAKVGNATIEDAKICKVGSLDPNKVKGKILFC--LLR 393
I G SLS N ++S+ +A G T + C SL+ K KGK+L C + R
Sbjct: 179 KITGESLSLFEMNAST-RIISASEAFAGYFTPYQSSYCLESSLNKTKTKGKVLVCRHVER 355
Query: 394 ELDGLVYAEEEAISGGSIGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATK 453
+ V + G +G++L ++ + +A ++P++ + G+ + SY+K T+
Sbjct: 356 STESKVAKSKIVKEAGGVGMILIDETDQD---VAIPFVIPSAIVGKKKGQKILSYLKTTR 526
Query: 454 TPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTG 513
PM+ + +AKT +G + AP +A+ SSRGPN + P ILKPDITAPG++IL A+ SP
Sbjct: 527 KPMSKILRAKTVIGAQSAPRVAAFSSRGPNALNPEILKPDITAPGLNILAAW----SP-- 688
Query: 514 LASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPI 573
+ N +NI SGTS++CPHV+ I L+K ++P+WSP+A KSAIMTT TI H+PI
Sbjct: 689 -VAGNM---FNILSGTSMACPHVTGIATLVKAVHPSWSPSAIKSAIMTTATILDKRHKPI 856
>TC88072 similar to PIR|T06580|T06580 subtilisin-like proteinase (EC
3.4.21.-) p69f - tomato, partial (27%)
Length = 1097
Score = 208 bits (529), Expect = 7e-54
Identities = 129/318 (40%), Positives = 176/318 (54%), Gaps = 6/318 (1%)
Frame = +2
Query: 427 AYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQ 486
A AH+LP ++Y G + SYIK+T P A + T +G AP + SSRGP+
Sbjct: 5 AIAHVLPAVEVSYAAGLTIKSYIKSTYNPTATLIFKGTIIGDSLAPSVVYFSSRGPSQES 184
Query: 487 PIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTI 546
P ILKPDI PGV+IL A+ ++ DN+ ++I SGTS+SCPH+S I AL+K+
Sbjct: 185 PGILKPDIIGPGVNILAAWAVSV-------DNKIPAFDIVSGTSMSCPHLSGIAALIKSS 343
Query: 547 YPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKEDATPFGYGAGHIQPELAMDPGLVYDLN 606
+P+WSPAA KSAIMTT PI DQ A F GAGH+ P A DPGLVYD+
Sbjct: 344 HPDWSPAAIKSAIMTTANTLNLGGIPILDQRLLPADIFATGAGHVNPFKANDPGLVYDIE 523
Query: 607 IVDYLNFLCAHGYNQTQMKMFSRKPYICP--KSYNMLDFNYPSITVPNLGKHFVQEVTRT 664
DY+ +LC GY+ ++++ + C KS NYPS ++ LG Q TRT
Sbjct: 524 PEDYVPYLCGLGYSDKEIEVIVQWKVKCSNVKSIPEAQLNYPSFSI-LLGSD-SQYYTRT 697
Query: 665 VTNVG-SPGTYRVQVNEPHGIFVLIKPRSLTFNEVGEKKTFKIIF-KVTKPTSSGYVFGH 722
+TNVG + TY+V++ P + + + P +TF EV EK +F + F K FG
Sbjct: 698 LTNVGFANSTYKVELEVPLALGMSVNPSEITFTEVNEKVSFSVEFIPQIKENRRNQTFGQ 877
Query: 723 --LLWSDGRHKVMSPLVV 738
L W +H V P+ V
Sbjct: 878 GSLTWVSDKHAVRVPISV 931
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,163,913
Number of Sequences: 36976
Number of extensions: 329765
Number of successful extensions: 1709
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 1526
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1540
length of query: 740
length of database: 9,014,727
effective HSP length: 103
effective length of query: 637
effective length of database: 5,206,199
effective search space: 3316348763
effective search space used: 3316348763
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147712.15


