

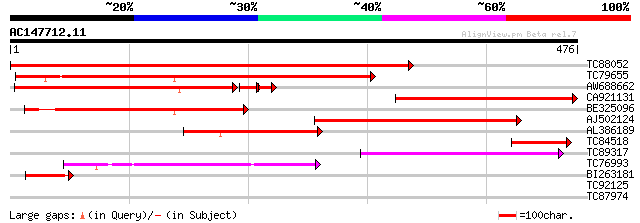
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147712.11 + phase: 0
(476 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88052 similar to GP|21554263|gb|AAM63338.1 putative uracil pho... 678 0.0
TC79655 similar to GP|10177966|dbj|BAB11349. uridine kinase-like... 435 e-122
AW688662 similar to GP|21554263|gb putative uracil phosphoribosy... 294 1e-85
CA921131 homologue to GP|21554263|gb putative uracil phosphoribo... 302 2e-82
BE325096 similar to GP|10177966|db uridine kinase-like protein {... 224 7e-59
AJ502124 similar to GP|11245466|gb cytosine deaminase-uracil pho... 191 7e-49
AL386189 similar to GP|8778301|gb| F14J16.5 {Arabidopsis thalian... 174 9e-44
TC84518 homologue to GP|21554263|gb|AAM63338.1 putative uracil p... 94 1e-19
TC89317 similar to SP|P93394|UPP_TOBAC Uracil phosphoribosyltran... 73 2e-13
TC76993 homologue to PIR|T06463|T06463 phosphoribulokinase (EC 2... 68 7e-12
BI263181 45 8e-05
TC92125 similar to GP|17065186|gb|AAL32747.1 putative protein {A... 29 3.5
TC87974 homologue to PIR|S71750|S71750 import intermediate-assoc... 28 7.7
>TC88052 similar to GP|21554263|gb|AAM63338.1 putative uracil phosphoribosyl
transferase {Arabidopsis thaliana}, partial (62%)
Length = 1155
Score = 678 bits (1750), Expect = 0.0
Identities = 337/339 (99%), Positives = 338/339 (99%)
Frame = +1
Query: 1 MGTAKSAVDLMGSSSEVHFSGFHMDGFEKREASIEQPTTSATDVYNQPFVIGVAGGAASG 60
MGTAKSAVDLMGSSSEVHFSGFHMDGFEKREASIEQPTTSATDVYNQPFVIGVAGGAASG
Sbjct: 139 MGTAKSAVDLMGSSSEVHFSGFHMDGFEKREASIEQPTTSATDVYNQPFVIGVAGGAASG 318
Query: 61 KKTVCDMIVQQLHDQRVVLVNQDSFYNNLTEEERARVQDYNFDHPDAFDTEELLRVMDKL 120
KKTVCDMIVQQLHDQRVVLVNQDSFYNNLTEEERARVQDYNFDHPDAFDTEELLRVMDKL
Sbjct: 319 KKTVCDMIVQQLHDQRVVLVNQDSFYNNLTEEERARVQDYNFDHPDAFDTEELLRVMDKL 498
Query: 121 KHGEAVDIPKYDFKSYKSDALRRVNPSDVIILEGILVFHDPRVRELMNMKIFVDTDADVR 180
KHGEAVDIPKYDFKSYKSDALRRVNPSDVIILEGILVFHDPRVRELMNMKIFVDTDADVR
Sbjct: 499 KHGEAVDIPKYDFKSYKSDALRRVNPSDVIILEGILVFHDPRVRELMNMKIFVDTDADVR 678
Query: 181 LARRIRRDTSEKDRDIGAILDQYSKFVKPAFDDFILPTKKYADIIIPRGGDNHVAVDLIV 240
LARRIRRDTSEKDRDIGAILDQYSKFVKPAFDDFILPTKKYADIIIPRGGDNHVAVDLIV
Sbjct: 679 LARRIRRDTSEKDRDIGAILDQYSKFVKPAFDDFILPTKKYADIIIPRGGDNHVAVDLIV 858
Query: 241 QHIRTKLGQHDLCKIYPNLYVIHSTFQIRGMHTLIRDAQITKHDFVFYSDRLIRLVVEHG 300
QHIRTKLGQHDLCKIYPNLYVIHSTFQIRGMHTLIRDAQ TKH+FVFYSDRLIRLVVEHG
Sbjct: 859 QHIRTKLGQHDLCKIYPNLYVIHSTFQIRGMHTLIRDAQKTKHEFVFYSDRLIRLVVEHG 1038
Query: 301 LGHLPFTEKQVITPTGSVYTGVDFCKRLCGVSVIRSGES 339
LGHLPFTEKQVITPTGSVYTGVDFCKRLCGVSVIRSGES
Sbjct: 1039LGHLPFTEKQVITPTGSVYTGVDFCKRLCGVSVIRSGES 1155
>TC79655 similar to GP|10177966|dbj|BAB11349. uridine kinase-like protein
{Arabidopsis thaliana}, partial (61%)
Length = 1015
Score = 435 bits (1119), Expect = e-122
Identities = 222/310 (71%), Positives = 251/310 (80%), Gaps = 8/310 (2%)
Frame = +1
Query: 6 SAVDL-MGSSSEVHFSGFHMDGFE-----KREASIEQPTTSATDVYNQPFVIGVAGGAAS 59
S++D M ++S HFSG +DG K SI T + + NQPFVIGV+GG AS
Sbjct: 88 SSIDYAMEAASGPHFSGLRLDGGRLSSSPKSSFSINSILTKDS-LLNQPFVIGVSGGTAS 264
Query: 60 GKKTVCDMIVQQLHDQRVVLVNQDSFYNNLTEEERARVQDYNFDHPDAFDTEELLRVMDK 119
GK TVCDMI+QQLHD RVVLVNQDSFY L EE V +YNFDHPDAFDTE+LL K
Sbjct: 265 GKTTVCDMIIQQLHDHRVVLVNQDSFYRGLKPEELEHVHEYNFDHPDAFDTEQLLECTKK 444
Query: 120 LKHGEAVDIPKYDFKSYK--SDALRRVNPSDVIILEGILVFHDPRVRELMNMKIFVDTDA 177
L G+ V IP YDFK ++ SD+ R+VN SDVIILEGILVFHD VR+LMNMKIFVDTDA
Sbjct: 445 LISGQRVKIPIYDFKKHQRSSDSFRQVNASDVIILEGILVFHDQDVRDLMNMKIFVDTDA 624
Query: 178 DVRLARRIRRDTSEKDRDIGAILDQYSKFVKPAFDDFILPTKKYADIIIPRGGDNHVAVD 237
DVRLARRIRRDT E+ RDI ++L+ Y+KFVKPAFDDF+LP+KKYAD+IIPRGGDNHVA+D
Sbjct: 625 DVRLARRIRRDTVERGRDINSVLEMYAKFVKPAFDDFVLPSKKYADVIIPRGGDNHVAID 804
Query: 238 LIVQHIRTKLGQHDLCKIYPNLYVIHSTFQIRGMHTLIRDAQITKHDFVFYSDRLIRLVV 297
LI QHIRTKLGQHDLCKIYPN+YVI STFQIRGMHTLIRD +I+KHDFVFYSDRLIRLVV
Sbjct: 805 LITQHIRTKLGQHDLCKIYPNVYVIQSTFQIRGMHTLIRDKEISKHDFVFYSDRLIRLVV 984
Query: 298 EHGLGHLPFT 307
EHGLGHLPFT
Sbjct: 985 EHGLGHLPFT 1014
>AW688662 similar to GP|21554263|gb putative uracil phosphoribosyl
transferase {Arabidopsis thaliana}, partial (34%)
Length = 685
Score = 294 bits (753), Expect(3) = 1e-85
Identities = 147/190 (77%), Positives = 164/190 (85%), Gaps = 3/190 (1%)
Frame = +2
Query: 5 KSAVDLMGSSSEVHFSGFHMD-GFEKREASIEQPTTSATDVYNQPFVIGVAGGAASGKKT 63
KS DLM +S+EVHFSGFHMD G E+R+A E+PTTSA D Y QPFVIGVAGG+ASGK
Sbjct: 14 KSVEDLMETSTEVHFSGFHMDNGLEQRKAGTEEPTTSAFDEYRQPFVIGVAGGSASGKTA 193
Query: 64 VCDMIVQQLHDQRVVLVNQDSFYNNLTEEERARVQDYNFDHPDAFDTEELLRVMDKLKHG 123
VCDMI+QQLHDQRVVLVNQDSFY+NLT+E+ RVQDYNFDHP+AFD+E LL VMDKLKH
Sbjct: 194 VCDMIIQQLHDQRVVLVNQDSFYHNLTKEQLTRVQDYNFDHPEAFDSERLLSVMDKLKHS 373
Query: 124 EAVDIPKYDFKSYKSDAL--RRVNPSDVIILEGILVFHDPRVRELMNMKIFVDTDADVRL 181
+AVDIPKYDFK YK+D RRVNP+DVIILEGILVFHDPRVR LMNMKIFVDTDADVRL
Sbjct: 374 QAVDIPKYDFKCYKNDVFPARRVNPADVIILEGILVFHDPRVRALMNMKIFVDTDADVRL 553
Query: 182 ARRIRRDTSE 191
A I+RDT +
Sbjct: 554 AXXIKRDTXD 583
Score = 33.5 bits (75), Expect(3) = 1e-85
Identities = 14/15 (93%), Positives = 14/15 (93%)
Frame = +1
Query: 210 AFDDFILPTKKYADI 224
AFDDF LPTKKYADI
Sbjct: 640 AFDDFFLPTKKYADI 684
Score = 28.9 bits (63), Expect(3) = 1e-85
Identities = 12/18 (66%), Positives = 14/18 (77%)
Frame = +3
Query: 194 RDIGAILDQYSKFVKPAF 211
R+I A+LDQYSKF P F
Sbjct: 591 RNIEAVLDQYSKFXXPGF 644
>CA921131 homologue to GP|21554263|gb putative uracil phosphoribosyl
transferase {Arabidopsis thaliana}, partial (30%)
Length = 654
Score = 302 bits (773), Expect = 2e-82
Identities = 150/152 (98%), Positives = 151/152 (98%)
Frame = -2
Query: 325 CKRLCGVSVIRSGESMENALRACCKGIKIGKILIHREGDNGQQLIYEKLPNDISERHVLL 384
CKRLCGVSVIRSG SM+NALRACCKGIKIGKILIHREGDNGQQLIYEKLPNDISERHVLL
Sbjct: 653 CKRLCGVSVIRSGTSMKNALRACCKGIKIGKILIHREGDNGQQLIYEKLPNDISERHVLL 474
Query: 385 LDPILGTGNSAVQAISLLIQKGVPESNIIFLNLISAPKGVHVVCKCFPRIKIVTSEIEIG 444
LDPILGTGNSAVQAISLLIQKGVPESNIIFLNLISAPKGVHVVCKCFPRIKIVTSEIEIG
Sbjct: 473 LDPILGTGNSAVQAISLLIQKGVPESNIIFLNLISAPKGVHVVCKCFPRIKIVTSEIEIG 294
Query: 445 LNEDFRVIPGMGEFGDRYFGTDDDEQVVVASQ 476
LNEDFRVIPGMGEFGDRYFGTDDDEQVVVASQ
Sbjct: 293 LNEDFRVIPGMGEFGDRYFGTDDDEQVVVASQ 198
>BE325096 similar to GP|10177966|db uridine kinase-like protein {Arabidopsis
thaliana}, partial (31%)
Length = 557
Score = 224 bits (570), Expect = 7e-59
Identities = 113/190 (59%), Positives = 141/190 (73%), Gaps = 2/190 (1%)
Frame = +3
Query: 13 SSSEVHFSGFHMDGFEKREASIEQPTTSATDVYNQPFVIGVAGGAASGKKTVCDMIVQQL 72
SSS HFSG H +++++ +N F+IGV+GG ASGK TVCDMI+QQL
Sbjct: 24 SSSAAHFSGCHPS------------SSTSSSSHNNVFLIGVSGGTASGKTTVCDMIIQQL 167
Query: 73 HDQRVVLVNQDSFYNNLTEEERARVQDYNFDHPDAFDTEELLRVMDKLKHGEAVDIPKYD 132
D RVVLVNQDSFY LT +E RV +YNFDHPDAFDTE+L+ + KLK G++V +P YD
Sbjct: 168 QDHRVVLVNQDSFYRGLTNDESKRVHEYNFDHPDAFDTEQLVETLIKLKSGQSVQVPVYD 347
Query: 133 FKSYK--SDALRRVNPSDVIILEGILVFHDPRVRELMNMKIFVDTDADVRLARRIRRDTS 190
FK ++ SD R+VN S+VIILEGILVFH+PRVR++MNMKIFVD D DVRL RRIRRDT
Sbjct: 348 FKLHQRASDRYRQVNASEVIILEGILVFHEPRVRDMMNMKIFVDADPDVRLGRRIRRDTV 527
Query: 191 EKDRDIGAIL 200
E+ RD+ ++L
Sbjct: 528 ERGRDVHSVL 557
>AJ502124 similar to GP|11245466|gb cytosine deaminase-uracil
phosphoribosyltransferase fusion protein {synthetic
construct}, partial (45%)
Length = 588
Score = 191 bits (484), Expect = 7e-49
Identities = 91/174 (52%), Positives = 125/174 (71%), Gaps = 1/174 (0%)
Frame = +2
Query: 257 PNLYVIHSTFQIRGMHTLIRDAQITKHDFVFYSDRLIRLVVEHGLGHLPFTEKQVITPTG 316
PN+ + T Q+ + T+IRD + DF+FYSDR+IRL+VE GL HLP EK V+TPT
Sbjct: 56 PNVVCLRETNQLEALLTMIRDKTTNRGDFIFYSDRIIRLLVEEGLNHLPVVEKTVVTPTS 235
Query: 317 SVYTGVDFCKRLCGVSVIRSGESMENALRACCKGIKIGKILIHREGDNGQ-QLIYEKLPN 375
+ + GV F ++CGVS++R+GE+ME LR CC+ ++IGKILI R+ + Q +L Y KLP
Sbjct: 236 NEFKGVGFKGKICGVSIMRAGEAMEQGLRDCCRSVRIGKILIQRDEETAQPKLYYAKLPP 415
Query: 376 DISERHVLLLDPILGTGNSAVQAISLLIQKGVPESNIIFLNLISAPKGVHVVCK 429
DI R+VLLLDP+L TG SA++AI +L+ GV E I+FLNLI+AP+G+ V K
Sbjct: 416 DIESRYVLLLDPMLATGGSAIKAIEVLLSHGVKEDKILFLNLIAAPEGIDAVTK 577
>AL386189 similar to GP|8778301|gb| F14J16.5 {Arabidopsis thaliana}, partial
(21%)
Length = 497
Score = 174 bits (440), Expect = 9e-44
Identities = 94/160 (58%), Positives = 106/160 (65%), Gaps = 44/160 (27%)
Frame = +2
Query: 147 SDVIILEGILVFHDPRVRELMNMKIFVDT------------------------------- 175
S+VIILEGILVFH+PRVR++MNMKIFVD
Sbjct: 14 SEVIILEGILVFHEPRVRDMMNMKIFVDAGLLIFQFCLSFQVVVPSITG*VG*NALNFVC 193
Query: 176 -------------DADVRLARRIRRDTSEKDRDIGAILDQYSKFVKPAFDDFILPTKKYA 222
D DVRL RRIRRDT E+ RD+ ++L+QY+KFVKPAFDDFILP+KKYA
Sbjct: 194 HREKEMFKVKE*NDPDVRLGRRIRRDTVERGRDVHSVLEQYAKFVKPAFDDFILPSKKYA 373
Query: 223 DIIIPRGGDNHVAVDLIVQHIRTKLGQHDLCKIYPNLYVI 262
DIIIPRGGDN VA+DLIVQHI TKLGQH LCKIYPNL VI
Sbjct: 374 DIIIPRGGDNCVAIDLIVQHIHTKLGQHWLCKIYPNLNVI 493
>TC84518 homologue to GP|21554263|gb|AAM63338.1 putative uracil
phosphoribosyl transferase {Arabidopsis thaliana},
partial (10%)
Length = 379
Score = 93.6 bits (231), Expect = 1e-19
Identities = 41/50 (82%), Positives = 48/50 (96%)
Frame = +2
Query: 422 KGVHVVCKCFPRIKIVTSEIEIGLNEDFRVIPGMGEFGDRYFGTDDDEQV 471
+G+HVVCK FPRIKIVTSEI+ GLNEDFRV+PGMGEFGDRYFGTDDD+++
Sbjct: 2 QGLHVVCKRFPRIKIVTSEIDNGLNEDFRVVPGMGEFGDRYFGTDDDDEL 151
>TC89317 similar to SP|P93394|UPP_TOBAC Uracil phosphoribosyltransferase (EC
2.4.2.9) (UMP pyrophosphorylase) (UPRTase). [Common
tobacco], partial (95%)
Length = 1129
Score = 73.2 bits (178), Expect = 2e-13
Identities = 48/174 (27%), Positives = 89/174 (50%), Gaps = 3/174 (1%)
Frame = +3
Query: 295 LVVEHGLGHLPFTEKQVITPTGSVYTG-VDFCKRLCGVSVIRSGESMENALRACCKGIKI 353
L+ E LP ++ +P G +D + + + ++R+G ++ + K
Sbjct: 369 LIYEASRDWLPTVSGEIQSPLGVASVEFIDPREPVAVIPILRAGLALAEHASSILPATKT 548
Query: 354 GKILIHREGDNGQQLIY-EKLPNDISE-RHVLLLDPILGTGNSAVQAISLLIQKGVPESN 411
+ I R + Q +Y KLP+ +E V ++DP+L TG + V A++LL ++GV
Sbjct: 549 YHLGITRNEETLQPTVYLNKLPDKFAEGSKVFVVDPMLATGGTIVAALNLLKERGVGNKQ 728
Query: 412 IIFLNLISAPKGVHVVCKCFPRIKIVTSEIEIGLNEDFRVIPGMGEFGDRYFGT 465
I ++ S+P + + + FP + + T I+ LN+ +IPG+G+ GDR +GT
Sbjct: 729 IKVISACSSPPALEKLSEQFPGLHVYTGIIDPVLNDQGFIIPGLGDAGDRSYGT 890
>TC76993 homologue to PIR|T06463|T06463 phosphoribulokinase (EC 2.7.1.19) -
garden pea (fragment), complete
Length = 1577
Score = 68.2 bits (165), Expect = 7e-12
Identities = 57/234 (24%), Positives = 105/234 (44%), Gaps = 18/234 (7%)
Frame = +2
Query: 46 NQPFVIGVAGGAASGKKTVCDMIVQQ------------------LHDQRVVLVNQDSFYN 87
+Q VIG+A + GK T + + D V+ D Y+
Sbjct: 299 SQTIVIGLAADSGCGKSTFMRRLTSVFGGAAEPPKGGNPDSNTLISDTTTVICLDD--YH 472
Query: 88 NLTEEERARVQDYNFDHPDAFDTEELLRVMDKLKHGEAVDIPKYDFKSYKSDALRRVNPS 147
+L R D P A D + + + +K G +V P Y+ + D + P
Sbjct: 473 SLDRTGRKEKGVTALD-PRANDFDLMYEQVKAIKEGVSVQKPIYNHVTGLLDPPELIKPP 649
Query: 148 DVIILEGILVFHDPRVRELMNMKIFVDTDADVRLARRIRRDTSEKDRDIGAILDQYSKFV 207
++++EG+ +D RVR+L++ I++D +V+ A +I+RD +E+ + +I +
Sbjct: 650 KILVIEGLHPMYDSRVRDLLDFSIYLDISNEVKFAWKIQRDMAERGHSLESIKASI-EAR 826
Query: 208 KPAFDDFILPTKKYADIIIPRGGDNHVAVDLIVQHIRTKLGQHDLCKIYPNLYV 261
KP F+ +I P K+YAD +I + D + +R +L Q + K + +Y+
Sbjct: 827 KPDFEAYIDPQKQYADAVIEVLPTQLIPDDNEGKILRVRLIQKEGVKYFSPVYL 988
>BI263181
Length = 628
Score = 44.7 bits (104), Expect = 8e-05
Identities = 23/40 (57%), Positives = 29/40 (72%)
Frame = +1
Query: 14 SSEVHFSGFHMDGFEKREASIEQPTTSATDVYNQPFVIGV 53
SS+VH + F+MDG E+R+A IEQPTTSA Y FV G+
Sbjct: 217 SSDVHLTIFYMDGLEQRKAEIEQPTTSAIGEYKH-FVKGL 333
>TC92125 similar to GP|17065186|gb|AAL32747.1 putative protein {Arabidopsis
thaliana}, partial (31%)
Length = 428
Score = 29.3 bits (64), Expect = 3.5
Identities = 21/71 (29%), Positives = 30/71 (41%), Gaps = 2/71 (2%)
Frame = +2
Query: 53 VAGGAASGKKTVCDMIVQQLHDQR--VVLVNQDSFYNNLTEEERARVQDYNFDHPDAFDT 110
V G A SGK T C + Q R + ++N D NFD+P A D
Sbjct: 191 VIGPAGSGKSTYCSSLYQHCETVRRSIHVMNLDPAAE-------------NFDYPVAMDV 331
Query: 111 EELLRVMDKLK 121
EL+ + D ++
Sbjct: 332 RELISLDDVME 364
>TC87974 homologue to PIR|S71750|S71750 import intermediate-associated 100K
protein precursor - garden pea, partial (68%)
Length = 2331
Score = 28.1 bits (61), Expect = 7.7
Identities = 25/80 (31%), Positives = 39/80 (48%), Gaps = 8/80 (10%)
Frame = +1
Query: 353 IGKILIHREGDNGQQLIYEKLPNDIS-----ERHVLLLDPILGTGNSAVQAISLLIQ--- 404
+G I G+ ++ +YEK+P D++ R V+ NS +QA++LL Q
Sbjct: 1651 VGDIFSSGTGEFDEEEVYEKIPLDLNINKEKARGVVRELAQSRLSNSLIQAVALLRQRNH 1830
Query: 405 KGVPESNIIFLNLISAPKGV 424
KGV S NL++ K V
Sbjct: 1831 KGVVSS---LNNLLACDKAV 1881
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.141 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,069,800
Number of Sequences: 36976
Number of extensions: 175437
Number of successful extensions: 765
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 755
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 758
length of query: 476
length of database: 9,014,727
effective HSP length: 100
effective length of query: 376
effective length of database: 5,317,127
effective search space: 1999239752
effective search space used: 1999239752
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147712.11


