

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147537.13 + phase: 0
(127 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
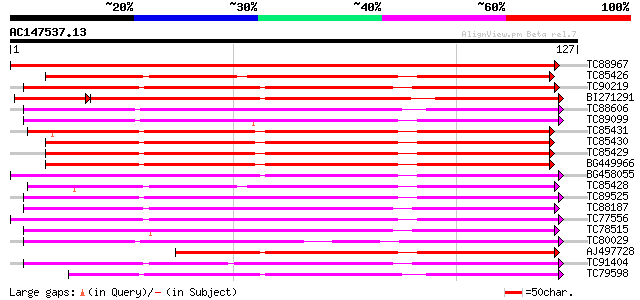
Score E
Sequences producing significant alignments: (bits) Value
TC88967 weakly similar to GP|18477856|emb|CAC86258. lipid transf... 258 4e-70
TC85426 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transf... 111 7e-26
TC90219 weakly similar to GP|20135538|gb|AAL25839.1 lipid transf... 108 7e-25
BI271291 similar to GP|6782436|gb|A lipid-transfer protein {Nico... 105 1e-24
TC88606 weakly similar to PIR|S71564|S71564 lipid transfer prote... 103 2e-23
TC89099 weakly similar to PIR|S71564|S71564 lipid transfer prote... 102 5e-23
TC85431 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transf... 100 2e-22
TC85430 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transf... 99 3e-22
TC85429 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transf... 99 4e-22
BG449966 similar to SP|O23758|NLTP Nonspecific lipid-transfer pr... 98 7e-22
BG458055 weakly similar to GP|16904376|gb| lipid transfer protei... 97 1e-21
TC85428 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transf... 96 5e-21
TC89525 95 8e-21
TC88187 weakly similar to PIR|T09790|T09790 lipid transfer prote... 93 2e-20
TC77556 weakly similar to SP|Q43766|NLT3_HORVU Nonspecific lipid... 92 5e-20
TC78515 91 2e-19
TC80029 similar to GP|6782436|gb|AAF28385.1| lipid-transfer prot... 90 2e-19
AJ497728 weakly similar to GP|20385449|gb| lipid-transfer protei... 88 8e-19
TC91404 87 1e-18
TC79598 similar to PIR|E84563|E84563 probable lipid transfer pro... 85 7e-18
>TC88967 weakly similar to GP|18477856|emb|CAC86258. lipid transfer protein
{Fragaria x ananassa}, partial (48%)
Length = 743
Score = 258 bits (659), Expect = 4e-70
Identities = 123/123 (100%), Positives = 123/123 (100%)
Frame = +1
Query: 1 MGYSTMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCN 60
MGYSTMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCN
Sbjct: 46 MGYSTMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCN 225
Query: 61 GIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNT 120
GIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNT
Sbjct: 226 GIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNT 405
Query: 121 DCN 123
DCN
Sbjct: 406 DCN 414
>TC85426 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transfer protein
precursor (LTP). [Chickpea Garbanzo] {Cicer arietinum},
complete
Length = 696
Score = 111 bits (278), Expect = 7e-26
Identities = 53/114 (46%), Positives = 75/114 (65%)
Frame = +1
Query: 9 RLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTM 68
++ C+ ++ + P AEAA+T CG V SL PC+ Y+ G P+A CC G++ LN+
Sbjct: 82 KVACVLLMMCIIVAPMAEAAIT-CGTVTGSLAPCIGYLKGGSG--PSAACCGGVKRLNSA 252
Query: 69 AQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
A TT DR+A C C+K+A + + LN N+AAGLP KCGVNIPY+IS +T+C
Sbjct: 253 ATTTPDRQAACNCLKSAAG----AISGLNPNIAAGLPGKCGVNIPYKISTSTNC 402
>TC90219 weakly similar to GP|20135538|gb|AAL25839.1 lipid transfer
precursor protein {Hevea brasiliensis}, partial (76%)
Length = 606
Score = 108 bits (269), Expect = 7e-25
Identities = 53/120 (44%), Positives = 77/120 (64%)
Frame = +2
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIR 63
++M ++ LA++CLV P A AA+ SCG + ++ PC+ Y+ G +VPA CCNGIR
Sbjct: 62 ASMFVKITFLAMICLVLGTPLANAAL-SCGQIQLTVAPCIGYLRTPGPSVPAP-CCNGIR 235
Query: 64 NLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
++ A+TT DR+ VC C+K+ S LNL AG P KCGVN+PY+++P+ DCN
Sbjct: 236 SVYYQAKTTADRQGVCRCLKSTT----LSLPGLNLPALAGAPAKCGVNVPYKVAPSIDCN 403
>BI271291 similar to GP|6782436|gb|A lipid-transfer protein {Nicotiana
glauca}, partial (23%)
Length = 575
Score = 105 bits (262), Expect(2) = 1e-24
Identities = 50/106 (47%), Positives = 71/106 (66%)
Frame = +3
Query: 19 VTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMAQTTNDRRAV 78
+ FG A SCG V+++LYPC+ YI N G +VPA CCNGIR +N A+ T+DR++V
Sbjct: 93 LVFGIPLANADLSCGQVLSTLYPCLGYIRNPGASVPAP-CCNGIRIVNDEAKNTSDRQSV 269
Query: 79 CTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCNR 124
C C+K+ + G +NL+ A LP CGVN+PY+I+P+ DCN+
Sbjct: 270 CRCLKSTIVLPG-----INLDALANLPTNCGVNLPYKITPDIDCNK 392
Score = 22.7 bits (47), Expect(2) = 1e-24
Identities = 8/17 (47%), Positives = 12/17 (70%)
Frame = +2
Query: 2 GYSTMATRLVCLAIVCL 18
G ++RL CLA++CL
Sbjct: 44 GQLNASSRLPCLAVICL 94
>TC88606 weakly similar to PIR|S71564|S71564 lipid transfer protein SDi-9
drought-induced - common sunflower, partial (23%)
Length = 604
Score = 103 bits (257), Expect = 2e-23
Identities = 53/121 (43%), Positives = 73/121 (59%)
Frame = +1
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIR 63
S+M T++ CLA++CLV P A +A SC V +L PC Y++ G P CCNG++
Sbjct: 46 SSMLTKVTCLAMICLVLSIPLANSA-PSCPEVQQTLAPCGPYLIQPGPPPPPEPCCNGVK 222
Query: 64 NLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
LN A+T DRR VC C+K+ V+ LNL A LP+ CGV+I Y ISP+ DC+
Sbjct: 223 TLNDQARTQQDRRDVCGCLKSIVAN-----PKLNLPAVASLPKDCGVDIGYVISPDMDCS 387
Query: 124 R 124
+
Sbjct: 388 K 390
>TC89099 weakly similar to PIR|S71564|S71564 lipid transfer protein SDi-9
drought-induced - common sunflower, partial (25%)
Length = 674
Score = 102 bits (253), Expect = 5e-23
Identities = 54/122 (44%), Positives = 73/122 (59%), Gaps = 1/122 (0%)
Frame = +1
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTV-PAAQCCNGI 62
S+M ++ CLA++CLV P A AA SC V +L PCV Y+ + G + P CCN +
Sbjct: 73 SSMFVKVTCLAMICLVLGIPLANAA-PSCPEVQQTLAPCVPYVTHPGPPISPPPPCCNAV 249
Query: 63 RNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
+ LN ++TT DRR VC C+K+ + LNL A LP+ CGV+I Y ISPN DC
Sbjct: 250 KTLNGQSKTTQDRRDVCGCLKSMMG----GIPGLNLPAIASLPKDCGVDIGYIISPNMDC 417
Query: 123 NR 124
N+
Sbjct: 418 NK 423
>TC85431 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transfer protein
precursor (LTP). [Chickpea Garbanzo] {Cicer arietinum},
complete
Length = 616
Score = 100 bits (248), Expect = 2e-22
Identities = 51/119 (42%), Positives = 73/119 (60%), Gaps = 1/119 (0%)
Frame = +3
Query: 5 TMAT-RLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIR 63
TMA ++ C+ ++ + P A+AA+ SCG V ++L PC+ Y+ G PA CC G++
Sbjct: 96 TMANMKIACVVLMMCMIVAPMADAAI-SCGTVTSALGPCIGYLKGGPGPSPA--CCGGVK 266
Query: 64 NLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
LN A TT DR+A C C+K A + + LN A+ LP KCGVNIPY+IS +T+C
Sbjct: 267 RLNGAAATTPDRQAACNCLKQAAG----AISGLNTAAASALPGKCGVNIPYKISTSTNC 431
>TC85430 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transfer protein
precursor (LTP). [Chickpea Garbanzo] {Cicer arietinum},
complete
Length = 646
Score = 99.4 bits (246), Expect = 3e-22
Identities = 48/114 (42%), Positives = 70/114 (61%)
Frame = +2
Query: 9 RLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTM 68
++ C+ ++ + P A+AA+ SCG V ++L PC+ Y+ G PA CC G++ LN
Sbjct: 62 KVACIVLMMCMIVAPMADAAI-SCGTVTSALGPCIGYLKGGPGPSPA--CCGGVKRLNGA 232
Query: 69 AQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
A TT DR+A C C+K A + + LN A+ LP KCGVNIPY+IS +T+C
Sbjct: 233 AATTPDRQAACNCLKQAAG----AISGLNTAAASALPGKCGVNIPYKISTSTNC 382
>TC85429 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transfer protein
precursor (LTP). [Chickpea Garbanzo] {Cicer arietinum},
complete
Length = 698
Score = 99.0 bits (245), Expect = 4e-22
Identities = 48/114 (42%), Positives = 70/114 (61%)
Frame = +3
Query: 9 RLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTM 68
++ C+ ++ + P A+AA+ SCG V ++L PC+ Y+ G PA CC G++ LN
Sbjct: 84 KVACVVLMMCMIVAPMADAAI-SCGTVTSALGPCIGYLKGGPGPSPA--CCGGVKRLNGA 254
Query: 69 AQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
A TT DR+A C C+K A + + LN A+ LP KCGVNIPY+IS +T+C
Sbjct: 255 AATTPDRQAACNCLKQAAG----AISGLNTAAASALPGKCGVNIPYKISTSTNC 404
>BG449966 similar to SP|O23758|NLTP Nonspecific lipid-transfer protein
precursor (LTP). [Chickpea Garbanzo] {Cicer arietinum},
complete
Length = 633
Score = 98.2 bits (243), Expect = 7e-22
Identities = 48/114 (42%), Positives = 69/114 (60%)
Frame = +3
Query: 9 RLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTM 68
++ C+ ++ + P A+AA+ SCG V ++L PC+ Y+ G PA CC G+ LN
Sbjct: 285 KVACIVLMMCMIVAPMADAAI-SCGTVTSALGPCIGYLKGGPGPSPA--CCGGVXRLNGA 455
Query: 69 AQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
A TT DR+A C C+K A + + LN A+ LP KCGVNIPY+IS +T+C
Sbjct: 456 AATTPDRQAACNCLKQAAG----AISGLNTAAASALPGKCGVNIPYKISTSTNC 605
>BG458055 weakly similar to GP|16904376|gb| lipid transfer protein {Setaria
italica}, partial (61%)
Length = 621
Score = 97.4 bits (241), Expect = 1e-21
Identities = 47/124 (37%), Positives = 69/124 (54%)
Frame = +2
Query: 1 MGYSTMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCN 60
M S + ++ CL ++ + G SC V T L PCV Y+ G VP CC+
Sbjct: 20 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQP-CCD 196
Query: 61 GIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNT 120
G++ +N A T +DR+A C CIK A S + LN+++ AGLP KCGV++PY + P+T
Sbjct: 197 GVKAINNQAVTKSDRQAACRCIKTATS----AIHGLNMDILAGLPSKCGVHLPYTLGPST 364
Query: 121 DCNR 124
C +
Sbjct: 365 TCEK 376
>TC85428 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transfer protein
precursor (LTP). [Chickpea Garbanzo] {Cicer arietinum},
partial (85%)
Length = 821
Score = 95.5 bits (236), Expect = 5e-21
Identities = 53/121 (43%), Positives = 69/121 (56%), Gaps = 2/121 (1%)
Frame = +2
Query: 5 TMATRLVCL--AIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGI 62
TMA+ V + AI+C+V AVT CG VV L PC++Y+ G P+A CC G+
Sbjct: 167 TMASLRVAIVAAIMCMVVVSAPMAEAVT-CGQVVGFLTPCITYLQGGPG--PSAACCGGV 337
Query: 63 RNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
+ LN A T R+ C C+K A + LN N AA LP KCGVNIPY+ S +T+C
Sbjct: 338 KKLNGAANTGPARKTACNCLKRAAG----NIARLNNNQAAALPGKCGVNIPYKFSTSTNC 505
Query: 123 N 123
N
Sbjct: 506 N 508
>TC89525
Length = 655
Score = 94.7 bits (234), Expect = 8e-21
Identities = 44/121 (36%), Positives = 71/121 (58%)
Frame = +2
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIR 63
S+M ++ CLA++CLV P A AA SC + +L PC+ Y+ + G+ P CCN ++
Sbjct: 44 SSMLVKVTCLAMICLVLGIPLANAAA-SCPEIEQTLTPCLEYVTHPGSPPPPEPCCNAVK 220
Query: 64 NLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
++ T DRR C+C+K+ + LNL AA + + CGV++ Y+ISP+ DC+
Sbjct: 221 AIHGQTHTPQDRRDCCSCVKSMIG----DIPGLNLPAAASMAKDCGVDLGYEISPDMDCS 388
Query: 124 R 124
+
Sbjct: 389 K 391
>TC88187 weakly similar to PIR|T09790|T09790 lipid transfer protein
precursor - upland cotton, partial (44%)
Length = 608
Score = 93.2 bits (230), Expect = 2e-20
Identities = 44/120 (36%), Positives = 68/120 (56%)
Frame = +1
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIR 63
S+M ++ C++++CLV P + A +T C V +LYPC Y G+ P A CC G++
Sbjct: 55 SSMLVKITCMSMLCLVLVIPLSNAGMT-CNEVTDTLYPCAGYATTPGDDPPPAGCCGGLK 231
Query: 64 NLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
++ A TT +R++VC C+K V +N + A LP KC V +PYQI + DC+
Sbjct: 232 DIKDKATTTPERQSVCECLKTNV----LRIPGVNPDTVAALPEKCDVPLPYQIKADFDCS 399
>TC77556 weakly similar to SP|Q43766|NLT3_HORVU Nonspecific lipid-transfer
protein 3 precursor (CW20) (CW-20) (CW-19). [Barley]
{Hordeum vulgare}, partial (28%)
Length = 662
Score = 92.0 bits (227), Expect = 5e-20
Identities = 46/124 (37%), Positives = 70/124 (56%)
Frame = +2
Query: 1 MGYSTMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCN 60
M S + ++ CLA++CLV P A A++T C V +L PC+ Y+ N P QCCN
Sbjct: 86 MASSMLVIKVTCLAMICLVLCIPLANASLT-CDEVKQNLTPCLPYVTNPHTLSPPDQCCN 262
Query: 61 GIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNT 120
G++ +N AQ DR+ VC C+K+ ++ LN +A+ LP CG+N I P+
Sbjct: 263 GVKTVNDNAQIKPDRQDVCRCLKSLLT----GVPGLNGTVASTLPSDCGINFRCPIGPDM 430
Query: 121 DCNR 124
DC++
Sbjct: 431 DCDK 442
>TC78515
Length = 639
Score = 90.5 bits (223), Expect = 2e-19
Identities = 46/121 (38%), Positives = 68/121 (56%), Gaps = 1/121 (0%)
Frame = +2
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVT-SCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGI 62
S+M ++ CL+++CL+ P A A C V S+ PCV YIM + P A CCNG+
Sbjct: 59 SSMLIKVTCLSVMCLLLAIPLANADPDPKCKNVAESIIPCVEYIMTPDASNPPAPCCNGM 238
Query: 63 RNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
+L Q +R+ C CIK+ + F +LNL A LP CGV++ +QI+P+ DC
Sbjct: 239 TSLAGQVQALPERQFACRCIKDGI----FDLPDLNLAALAALPNNCGVDLRFQITPDMDC 406
Query: 123 N 123
+
Sbjct: 407 D 409
>TC80029 similar to GP|6782436|gb|AAF28385.1| lipid-transfer protein
{Nicotiana glauca}, partial (23%)
Length = 539
Score = 90.1 bits (222), Expect = 2e-19
Identities = 46/121 (38%), Positives = 71/121 (58%)
Frame = +3
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIR 63
S+M ++ CL+++CLV P A A C VV ++ PC+ YI G + A CCNG++
Sbjct: 48 SSMLIKVTCLSVMCLVLAIPLANAG-PYCRDVVETILPCIEYITTPGASTLPAPCCNGMK 224
Query: 64 NLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
+LN + + VC C+K ++ F LNL A LP+ CGVN+PYQI+P+ +C+
Sbjct: 225 SLN------GEPQYVCRCLK----ETFFVLPGLNLAALAALPKNCGVNLPYQITPDMNCD 374
Query: 124 R 124
+
Sbjct: 375 K 377
>AJ497728 weakly similar to GP|20385449|gb| lipid-transfer protein {Citrus
sinensis}, partial (55%)
Length = 560
Score = 88.2 bits (217), Expect = 8e-19
Identities = 41/86 (47%), Positives = 57/86 (65%)
Frame = -1
Query: 38 SLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLN 97
++ PC+ Y+ N G +VPA CCNG+R LN A+TT +R++VC C+K S + LN
Sbjct: 554 TVVPCLGYLRNPGPSVPAP-CCNGLRGLNNQAKTTPERQSVCRCLKTTAQ----SLSGLN 390
Query: 98 LNLAAGLPRKCGVNIPYQISPNTDCN 123
+ A LP+KCGVN+PY+IS DCN
Sbjct: 389 VPALATLPKKCGVNLPYKISTAIDCN 312
>TC91404
Length = 595
Score = 87.4 bits (215), Expect = 1e-18
Identities = 45/121 (37%), Positives = 68/121 (56%)
Frame = +3
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIR 63
S+M ++ CLA++CLV P A A +T C +L C+SY+ N P CC+ +
Sbjct: 63 SSMLVKVTCLAMICLVLGIPLANAVIT-CPDTDITLTACLSYVAYP-NPPPPQPCCDAVL 236
Query: 64 NLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
++ A+T DR+AVC+C+K + L+L A LP+ CG NI Y+ISP DC+
Sbjct: 237 DVTAQARTREDRQAVCSCLKGLM----IGIPGLDLTALAALPKVCGANIGYEISPGMDCS 404
Query: 124 R 124
+
Sbjct: 405 K 407
>TC79598 similar to PIR|E84563|E84563 probable lipid transfer protein
[imported] - Arabidopsis thaliana, partial (58%)
Length = 695
Score = 85.1 bits (209), Expect = 7e-18
Identities = 43/111 (38%), Positives = 63/111 (56%)
Frame = +1
Query: 14 AIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMAQTTN 73
A+V L+ +EAA+ SC V+ L PCVSY+++G PAA CC+G + L + T+
Sbjct: 34 ALVLLIFVASNSEAAI-SCSDVIKDLKPCVSYLVSGSGQPPAA-CCSGAKALASAVSTSE 207
Query: 74 DRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCNR 124
D++A C CIK+ +N LA L CG+N P ISP+ DC++
Sbjct: 208 DKKAACNCIKSTSKS-----IKINSQLAQALAGNCGINTPITISPDADCSK 345
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.134 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,720,305
Number of Sequences: 36976
Number of extensions: 63589
Number of successful extensions: 387
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 338
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 342
length of query: 127
length of database: 9,014,727
effective HSP length: 85
effective length of query: 42
effective length of database: 5,871,767
effective search space: 246614214
effective search space used: 246614214
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 52 (24.6 bits)
Medicago: description of AC147537.13


