

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147537.12 + phase: 0
(125 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
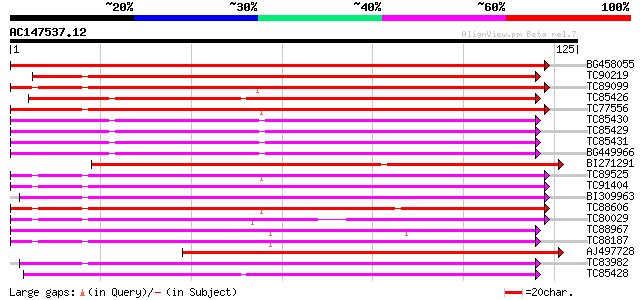
Score E
Sequences producing significant alignments: (bits) Value
BG458055 weakly similar to GP|16904376|gb| lipid transfer protei... 249 2e-67
TC90219 weakly similar to GP|20135538|gb|AAL25839.1 lipid transf... 116 3e-27
TC89099 weakly similar to PIR|S71564|S71564 lipid transfer prote... 111 6e-26
TC85426 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transf... 111 8e-26
TC77556 weakly similar to SP|Q43766|NLT3_HORVU Nonspecific lipid... 109 3e-25
TC85430 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transf... 108 4e-25
TC85429 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transf... 108 5e-25
TC85431 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transf... 108 7e-25
BG449966 similar to SP|O23758|NLTP Nonspecific lipid-transfer pr... 107 2e-24
BI271291 similar to GP|6782436|gb|A lipid-transfer protein {Nico... 105 4e-24
TC89525 105 4e-24
TC91404 100 1e-22
BI309963 100 3e-22
TC88606 weakly similar to PIR|S71564|S71564 lipid transfer prote... 99 4e-22
TC80029 similar to GP|6782436|gb|AAF28385.1| lipid-transfer prot... 98 7e-22
TC88967 weakly similar to GP|18477856|emb|CAC86258. lipid transf... 97 2e-21
TC88187 weakly similar to PIR|T09790|T09790 lipid transfer prote... 94 1e-20
AJ497728 weakly similar to GP|20385449|gb| lipid-transfer protei... 93 3e-20
TC83982 88 1e-18
TC85428 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transf... 87 2e-18
>BG458055 weakly similar to GP|16904376|gb| lipid transfer protein {Setaria
italica}, partial (61%)
Length = 621
Score = 249 bits (636), Expect = 2e-67
Identities = 119/119 (100%), Positives = 119/119 (100%)
Frame = +2
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDG 60
MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDG
Sbjct: 20 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDG 199
Query: 61 VKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCEK 119
VKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCEK
Sbjct: 200 VKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCEK 376
>TC90219 weakly similar to GP|20135538|gb|AAL25839.1 lipid transfer
precursor protein {Hevea brasiliensis}, partial (76%)
Length = 606
Score = 116 bits (290), Expect = 3e-27
Identities = 52/112 (46%), Positives = 76/112 (67%)
Frame = +2
Query: 6 VLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVKAIN 65
+ VK+T L M+ C+VLG P ALSC Q++ + PC+ Y+ G VP PCC+G++++
Sbjct: 68 MFVKITFLAMI-CLVLGTPLANAALSCGQIQLTVAPCIGYLRTPGPSVPAPCCNGIRSVY 244
Query: 66 NQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
QA T +DRQ CRC+K+ T ++ GLN+ LAG P+KCGV++PY + PS C
Sbjct: 245 YQAKTTADRQGVCRCLKSTTLSLPGLNLPALAGAPAKCGVNVPYKVAPSIDC 400
>TC89099 weakly similar to PIR|S71564|S71564 lipid transfer protein SDi-9
drought-induced - common sunflower, partial (25%)
Length = 674
Score = 111 bits (278), Expect = 6e-26
Identities = 57/121 (47%), Positives = 75/121 (61%), Gaps = 2/121 (1%)
Frame = +1
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYV--PQPCC 58
M+SSM VKVTCL M+ C+VLG+P A SC +V+ L PCVPYVT G + P PCC
Sbjct: 67 MASSM-FVKVTCLAMI-CLVLGIPLANAAPSCPEVQQTLAPCVPYVTHPGPPISPPPPCC 240
Query: 59 DGVKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCE 118
+ VK +N Q+ T DR+ C C+K+ I GLN+ +A LP CGV + Y + P+ C
Sbjct: 241 NAVKTLNGQSKTTQDRRDVCGCLKSMMGGIPGLNLPAIASLPKDCGVDIGYIISPNMDCN 420
Query: 119 K 119
K
Sbjct: 421 K 423
>TC85426 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transfer protein
precursor (LTP). [Chickpea Garbanzo] {Cicer arietinum},
complete
Length = 696
Score = 111 bits (277), Expect = 8e-26
Identities = 53/113 (46%), Positives = 71/113 (61%)
Frame = +1
Query: 5 MVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVKAI 64
M +KV C+ +MMC+++ P A++C V L PC+ Y+ G G CC GVK +
Sbjct: 70 MASMKVACVLLMMCIIVA-PMAEAAITCGTVTGSLAPCIGYLKGGSG-PSAACCGGVKRL 243
Query: 65 NNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
N+ A T DRQAAC C+K+A AI GLN +I AGLP KCGV++PY + ST C
Sbjct: 244 NSAATTTPDRQAACNCLKSAAGAISGLNPNIAAGLPGKCGVNIPYKISTSTNC 402
>TC77556 weakly similar to SP|Q43766|NLT3_HORVU Nonspecific lipid-transfer
protein 3 precursor (CW20) (CW-20) (CW-19). [Barley]
{Hordeum vulgare}, partial (28%)
Length = 662
Score = 109 bits (272), Expect = 3e-25
Identities = 52/120 (43%), Positives = 77/120 (63%), Gaps = 1/120 (0%)
Frame = +2
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVP-QPCCD 59
M+SSM+++KVTCL M+ C+VL +P +L+C +V+ L PC+PYVT P CC+
Sbjct: 86 MASSMLVIKVTCLAMI-CLVLCIPLANASLTCDEVKQNLTPCLPYVTNPHTLSPPDQCCN 262
Query: 60 GVKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCEK 119
GVK +N+ A K DRQ CRC+K+ + + GLN + + LPS CG++ +GP C+K
Sbjct: 263 GVKTVNDNAQIKPDRQDVCRCLKSLLTGVPGLNGTVASTLPSDCGINFRCPIGPDMDCDK 442
>TC85430 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transfer protein
precursor (LTP). [Chickpea Garbanzo] {Cicer arietinum},
complete
Length = 646
Score = 108 bits (271), Expect = 4e-25
Identities = 53/117 (45%), Positives = 69/117 (58%)
Frame = +2
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDG 60
+ M +KV C+ +MMCM++ P A+SC V + L PC+ Y+ G G P CC G
Sbjct: 38 LRKKMASMKVACIVLMMCMIVA-PMADAAISCGTVTSALGPCIGYLKGGPGPSPA-CCGG 211
Query: 61 VKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
VK +N A T DRQAAC C+K A AI GLN + LP KCGV++PY + ST C
Sbjct: 212 VKRLNGAAATTPDRQAACNCLKQAAGAISGLNTAAASALPGKCGVNIPYKISTSTNC 382
>TC85429 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transfer protein
precursor (LTP). [Chickpea Garbanzo] {Cicer arietinum},
complete
Length = 698
Score = 108 bits (270), Expect = 5e-25
Identities = 53/117 (45%), Positives = 69/117 (58%)
Frame = +3
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDG 60
+ M +KV C+ +MMCM++ P A+SC V + L PC+ Y+ G G P CC G
Sbjct: 60 LRKKMASMKVACVVLMMCMIVA-PMADAAISCGTVTSALGPCIGYLKGGPGPSPA-CCGG 233
Query: 61 VKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
VK +N A T DRQAAC C+K A AI GLN + LP KCGV++PY + ST C
Sbjct: 234 VKRLNGAAATTPDRQAACNCLKQAAGAISGLNTAAASALPGKCGVNIPYKISTSTNC 404
>TC85431 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transfer protein
precursor (LTP). [Chickpea Garbanzo] {Cicer arietinum},
complete
Length = 616
Score = 108 bits (269), Expect = 7e-25
Identities = 52/117 (44%), Positives = 70/117 (59%)
Frame = +3
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDG 60
+ +M +K+ C+ +MMCM++ P A+SC V + L PC+ Y+ G G P CC G
Sbjct: 87 LRKTMANMKIACVVLMMCMIVA-PMADAAISCGTVTSALGPCIGYLKGGPGPSPA-CCGG 260
Query: 61 VKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
VK +N A T DRQAAC C+K A AI GLN + LP KCGV++PY + ST C
Sbjct: 261 VKRLNGAAATTPDRQAACNCLKQAAGAISGLNTAAASALPGKCGVNIPYKISTSTNC 431
>BG449966 similar to SP|O23758|NLTP Nonspecific lipid-transfer protein
precursor (LTP). [Chickpea Garbanzo] {Cicer arietinum},
complete
Length = 633
Score = 107 bits (266), Expect = 2e-24
Identities = 52/117 (44%), Positives = 68/117 (57%)
Frame = +3
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDG 60
+ M +KV C+ +MMCM++ P A+SC V + L PC+ Y+ G G P CC G
Sbjct: 261 LXKKMASMKVACIVLMMCMIVA-PMADAAISCGTVTSALGPCIGYLKGGPGPSPA-CCGG 434
Query: 61 VKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
V +N A T DRQAAC C+K A AI GLN + LP KCGV++PY + ST C
Sbjct: 435 VXRLNGAAATTPDRQAACNCLKQAAGAISGLNTAAASALPGKCGVNIPYKISTSTNC 605
>BI271291 similar to GP|6782436|gb|A lipid-transfer protein {Nicotiana
glauca}, partial (23%)
Length = 575
Score = 105 bits (263), Expect = 4e-24
Identities = 47/104 (45%), Positives = 67/104 (64%)
Frame = +3
Query: 19 MVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVKAINNQAVTKSDRQAAC 78
+V G+P LSC QV + L PC+ Y+ G VP PCC+G++ +N++A SDRQ+ C
Sbjct: 93 LVFGIPLANADLSCGQVLSTLYPCLGYIRNPGASVPAPCCNGIRIVNDEAKNTSDRQSVC 272
Query: 79 RCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCEKYVY 122
RC+K +T + G+N+D LA LP+ CGV+LPY + P C K Y
Sbjct: 273 RCLK-STIVLPGINLDALANLPTNCGVNLPYKITPDIDCNKIPY 401
>TC89525
Length = 655
Score = 105 bits (263), Expect = 4e-24
Identities = 56/120 (46%), Positives = 72/120 (59%), Gaps = 1/120 (0%)
Frame = +2
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVP-QPCCD 59
M+SSM LVKVTCL M+ C+VLG+P A SC ++E L PC+ YVT G P +PCC+
Sbjct: 38 MASSM-LVKVTCLAMI-CLVLGIPLANAAASCPEIEQTLTPCLEYVTHPGSPPPPEPCCN 211
Query: 60 GVKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCEK 119
VKAI+ Q T DR+ C C+K+ I GLN+ A + CGV L Y + P C K
Sbjct: 212 AVKAIHGQTHTPQDRRDCCSCVKSMIGDIPGLNLPAAASMAKDCGVDLGYEISPDMDCSK 391
>TC91404
Length = 595
Score = 100 bits (250), Expect = 1e-22
Identities = 52/119 (43%), Positives = 68/119 (56%)
Frame = +3
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDG 60
M+SSM LVKVTCL M+ C+VLG+P ++C + L C+ YV PQPCCD
Sbjct: 57 MASSM-LVKVTCLAMI-CLVLGIPLANAVITCPDTDITLTACLSYVAYPNPPPPQPCCDA 230
Query: 61 VKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCEK 119
V + QA T+ DRQA C C+K I GL++ LA LP CG ++ Y + P C K
Sbjct: 231 VLDVTAQARTREDRQAVCSCLKGLMIGIPGLDLTALAALPKVCGANIGYEISPGMDCSK 407
>BI309963
Length = 529
Score = 99.8 bits (247), Expect = 3e-22
Identities = 48/117 (41%), Positives = 66/117 (56%)
Frame = +1
Query: 3 SSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVK 62
+S +LVKVTCL + C+VLG+P ++C LM C+PYV PQ CC V
Sbjct: 1 ASSMLVKVTCLATI-CLVLGIPLANAVITCRDAAITLMGCLPYVAHPTPSPPQNCCAAVL 177
Query: 63 AINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCEK 119
+ QA+T+ DRQA C C+K + I GL++ LA LP CG ++ Y + P C K
Sbjct: 178 DVTGQAITREDRQAVCSCLKGLMNGIPGLDLTALASLPKVCGANIGYEISPDMDCSK 348
>TC88606 weakly similar to PIR|S71564|S71564 lipid transfer protein SDi-9
drought-induced - common sunflower, partial (23%)
Length = 604
Score = 99.0 bits (245), Expect = 4e-22
Identities = 54/120 (45%), Positives = 73/120 (60%), Gaps = 1/120 (0%)
Frame = +1
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVP-QPCCD 59
M+SSM L KVTCL M+ C+VL +P A SC +V+ L PC PY+ G P +PCC+
Sbjct: 40 MASSM-LTKVTCLAMI-CLVLSIPLANSAPSCPEVQQTLAPCGPYLIQPGPPPPPEPCCN 213
Query: 60 GVKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCEK 119
GVK +N+QA T+ DR+ C C+K+ A LN+ +A LP CGV + Y + P C K
Sbjct: 214 GVKTLNDQARTQQDRRDVCGCLKSIV-ANPKLNLPAVASLPKDCGVDIGYVISPDMDCSK 390
>TC80029 similar to GP|6782436|gb|AAF28385.1| lipid-transfer protein
{Nicotiana glauca}, partial (23%)
Length = 539
Score = 98.2 bits (243), Expect = 7e-22
Identities = 50/120 (41%), Positives = 72/120 (59%), Gaps = 1/120 (0%)
Frame = +3
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGY-VPQPCCD 59
M+SSM L+KVTCL++M C+VL +P C V ++PC+ Y+T G +P PCC+
Sbjct: 42 MASSM-LIKVTCLSVM-CLVLAIPLANAGPYCRDVVETILPCIEYITTPGASTLPAPCCN 215
Query: 60 GVKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCEK 119
G+K++N + Q CRC+K + GLN+ LA LP CGV+LPY + P C+K
Sbjct: 216 GMKSLNGEP------QYVCRCLKETFFVLPGLNLAALAALPKNCGVNLPYQITPDMNCDK 377
>TC88967 weakly similar to GP|18477856|emb|CAC86258. lipid transfer protein
{Fragaria x ananassa}, partial (48%)
Length = 743
Score = 96.7 bits (239), Expect = 2e-21
Identities = 47/122 (38%), Positives = 68/122 (55%), Gaps = 5/122 (4%)
Frame = +1
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQP-CCD 59
M S + ++ CL ++ + G SC V T L PCV Y+ G VP CC+
Sbjct: 46 MGYSTMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCN 225
Query: 60 GVKAINNQAVTKSDRQAACRCIKTATS----AIHGLNMDILAGLPSKCGVHLPYTLGPST 115
G++ +N A T +DR+A C CIK A S + LN+++ AGLP KCGV++PY + P+T
Sbjct: 226 GIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNT 405
Query: 116 TC 117
C
Sbjct: 406 DC 411
>TC88187 weakly similar to PIR|T09790|T09790 lipid transfer protein
precursor - upland cotton, partial (44%)
Length = 608
Score = 94.4 bits (233), Expect = 1e-20
Identities = 50/118 (42%), Positives = 70/118 (58%), Gaps = 1/118 (0%)
Frame = +1
Query: 1 MSSSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQP-CCD 59
M+SSM LVK+TC++M+ C+VL +P + ++C +V L PC Y T G P CC
Sbjct: 49 MASSM-LVKITCMSML-CLVLVIPLSNAGMTCNEVTDTLYPCAGYATTPGDDPPPAGCCG 222
Query: 60 GVKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
G+K I ++A T +RQ+ C C+KT I G+N D +A LP KC V LPY + C
Sbjct: 223 GLKDIKDKATTTPERQSVCECLKTNVLRIPGVNPDTVAALPEKCDVPLPYQIKADFDC 396
>AJ497728 weakly similar to GP|20385449|gb| lipid-transfer protein {Citrus
sinensis}, partial (55%)
Length = 560
Score = 92.8 bits (229), Expect = 3e-20
Identities = 38/84 (45%), Positives = 55/84 (65%)
Frame = -1
Query: 39 LMPCVPYVTGNGGYVPQPCCDGVKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAG 98
++PC+ Y+ G VP PCC+G++ +NNQA T +RQ+ CRC+KT ++ GLN+ LA
Sbjct: 551 VVPCLGYLRNPGPSVPAPCCNGLRGLNNQAKTTPERQSVCRCLKTTAQSLSGLNVPALAT 372
Query: 99 LPSKCGVHLPYTLGPSTTCEKYVY 122
LP KCGV+LPY + + C Y
Sbjct: 371 LPKKCGVNLPYKISTAIDCNTVKY 300
>TC83982
Length = 608
Score = 87.8 bits (216), Expect = 1e-18
Identities = 41/115 (35%), Positives = 64/115 (55%)
Frame = +2
Query: 3 SSMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVK 62
+S +LVKVTCL M+ C+VL +P ++C + L C+PYV + P CC V
Sbjct: 71 ASSILVKVTCLAMI-CLVLSIPLANAVITCPDADITLKSCLPYVAHPTQWPPLECCTAVL 247
Query: 63 AINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
+ +AVT+ DRQA C+C+ + I GL++ A +P C ++ Y + P+ C
Sbjct: 248 GLTARAVTREDRQAVCKCLLGLMNGIPGLDLTAFAAVPILCAANIGYIIRPNMDC 412
>TC85428 similar to SP|O23758|NLTP_CICAR Nonspecific lipid-transfer protein
precursor (LTP). [Chickpea Garbanzo] {Cicer arietinum},
partial (85%)
Length = 821
Score = 87.0 bits (214), Expect = 2e-18
Identities = 45/114 (39%), Positives = 62/114 (53%)
Frame = +2
Query: 4 SMVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVKA 63
+M ++V + +MCMV+ +A++C QV L PC+ Y+ G G CC GVK
Sbjct: 167 TMASLRVAIVAAIMCMVVVSAPMAEAVTCGQVVGFLTPCITYLQGGPG-PSAACCGGVKK 343
Query: 64 INNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
+N A T R+ AC C+K A I LN + A LP KCGV++PY ST C
Sbjct: 344 LNGAANTGPARKTACNCLKRAAGNIARLNNNQAAALPGKCGVNIPYKFSTSTNC 505
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.135 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,691,897
Number of Sequences: 36976
Number of extensions: 63448
Number of successful extensions: 384
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 345
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 347
length of query: 125
length of database: 9,014,727
effective HSP length: 84
effective length of query: 41
effective length of database: 5,908,743
effective search space: 242258463
effective search space used: 242258463
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 52 (24.6 bits)
Medicago: description of AC147537.12


