

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147536.1 + phase: 0
(648 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
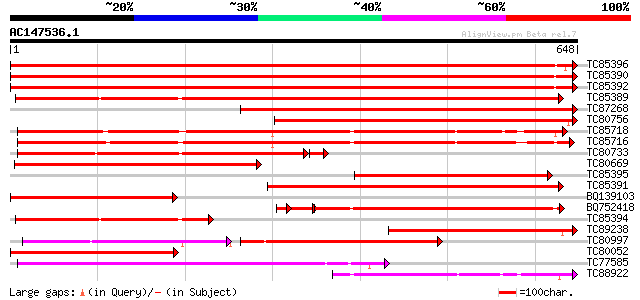
Score E
Sequences producing significant alignments: (bits) Value
TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protei... 1200 0.0
TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 ... 1195 0.0
TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chape... 1191 0.0
TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protei... 809 0.0
TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chape... 706 0.0
TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 591 e-169
TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa prot... 560 e-160
TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chape... 546 e-156
TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa prot... 525 e-155
TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 507 e-144
TC85395 homologue to GP|6911553|emb|CAB72130.1 heat shock protei... 448 e-126
TC85391 homologue to PIR|T06598|T06598 dnaK-type molecular chape... 434 e-122
BQ139103 homologue to GP|2655420|gb| heat shock cognate protein ... 366 e-101
BQ752418 homologue to SP|P78695|GR78 78 kDa glucose-regulated pr... 336 e-101
TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic retic... 313 2e-85
TC89238 similar to SP|Q01877|HS71_PUCGR Heat shock protein HSS1.... 300 2e-81
TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [im... 181 6e-76
TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chapero... 276 2e-74
TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock... 270 1e-72
TC88922 homologue to SP|Q02028|HS7S_PEA Stromal 70 kDa heat shoc... 221 5e-58
>TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protein 70
{Cucumis sativus}, complete
Length = 2320
Score = 1200 bits (3105), Expect = 0.0
Identities = 609/651 (93%), Positives = 628/651 (95%), Gaps = 3/651 (0%)
Frame = +2
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN
Sbjct: 107 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 286
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
QVAMNP NTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGE+K FA+
Sbjct: 287 QVAMNPTNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEEKLFAS 466
Query: 121 EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 180
EEISSMVLIKMREIAEAYLG T+KNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP
Sbjct: 467 EEISSMVLIKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 646
Query: 181 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 240
TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD
Sbjct: 647 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 826
Query: 241 NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVDFY 300
NRMV HFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVDFY
Sbjct: 827 NRMVTHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVDFY 1006
Query: 301 TTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQDFF 360
TTITRARFEELNMDLFRKCMEPVEKCLRDAKMDK++VHDVVLVGGSTRIPKVQQLLQDFF
Sbjct: 1007TTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLLQDFF 1186
Query: 361 NGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLI 420
NGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLS GLETAGGVMTVLI
Sbjct: 1187NGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSQGLETAGGVMTVLI 1366
Query: 421 PRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERARTRDNNLLGKFELSGIPPAPRGVPQI 480
PRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGER RT+DNNLLGKFELSGIPPAPRGVPQI
Sbjct: 1367PRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTKDNNLLGKFELSGIPPAPRGVPQI 1546
Query: 481 TVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVEEAEKYKAEDEEHKK 540
TVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMV+EAEKYK+EDEEHKK
Sbjct: 1547TVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDEEHKK 1726
Query: 541 KVDAKNALENYAYNMRNTIKDEKIGSKLSPEDKKKIDDAIDAAIQWLDSNQLAEADEFED 600
KV+AKN+LENYAYNMRNTIKDEKI SKLS DKK+I+DAI+ AIQWLD+NQLAEADEFED
Sbjct: 1727KVEAKNSLENYAYNMRNTIKDEKISSKLSGGDKKQIEDAIEGAIQWLDANQLAEADEFED 1906
Query: 601 KMKELESLCNPIIAKMYQGGGAPDMGGGMDDEV---PPAAGGAGPKIEEVD 648
KMKELE++CNPIIAKMYQGG G +DD+ P +GGAGPKIEEVD
Sbjct: 1907KMKELETICNPIIAKMYQGGAGE--GPEVDDDAAPPPSGSGGAGPKIEEVD 2053
>TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 kDa protein
2. [Tomato] {Lycopersicon esculentum}, complete
Length = 2278
Score = 1195 bits (3092), Expect = 0.0
Identities = 602/650 (92%), Positives = 623/650 (95%), Gaps = 2/650 (0%)
Frame = +1
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTD+ERLIGDAAKN
Sbjct: 136 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKN 315
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
QVAMNP NTVFDAKRLIGRR D SVQSDMKLWPFKVI GP +KPMI VNYK E+KQF+A
Sbjct: 316 QVAMNPTNTVFDAKRLIGRRFGDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQFSA 495
Query: 121 EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 180
EEISSMVL+KM+EIAEAYLG+T+KNAVVTVPAYFNDSQRQATKDAGVI+GLNVLRIINEP
Sbjct: 496 EEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVLRIINEP 675
Query: 181 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 240
TAAAIAYGLDKKATS GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD
Sbjct: 676 TAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 855
Query: 241 NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVDFY 300
NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEG+DFY
Sbjct: 856 NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGIDFY 1035
Query: 301 TTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQDFF 360
TTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQDFF
Sbjct: 1036TTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQDFF 1215
Query: 361 NGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLI 420
NGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLI
Sbjct: 1216NGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLI 1395
Query: 421 PRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERARTRDNNLLGKFELSGIPPAPRGVPQI 480
PRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGER RTRDNNLLGKFELSGIPPAPRGVPQI
Sbjct: 1396PRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRGVPQI 1575
Query: 481 TVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVEEAEKYKAEDEEHKK 540
TVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMV+EAEKYK+EDEEHKK
Sbjct: 1576TVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDEEHKK 1755
Query: 541 KVDAKNALENYAYNMRNTIKDEKIGSKLSPEDKKKIDDAIDAAIQWLDSNQLAEADEFED 600
KV+AKNALENYAYNMRNTIKD+KI SKLS +DKKKI+DAI+ AIQWLD NQL EADEFED
Sbjct: 1756KVEAKNALENYAYNMRNTIKDDKIASKLSADDKKKIEDAIEGAIQWLDGNQLGEADEFED 1935
Query: 601 KMKELESLCNPIIAKMYQGGGAPDMGGGMDDEVPPA--AGGAGPKIEEVD 648
KMKELE +CNPIIA+MYQG G D GG MD++ P A GAGPKIEEVD
Sbjct: 1936KMKELEGICNPIIARMYQGAGG-DAGGAMDEDGPTAGSGSGAGPKIEEVD 2082
>TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chaperone
hsc70-3 - tomato, complete
Length = 2295
Score = 1191 bits (3080), Expect = 0.0
Identities = 599/650 (92%), Positives = 624/650 (95%), Gaps = 2/650 (0%)
Frame = +1
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTD+ERLIGDAAKN
Sbjct: 130 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKN 309
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
QVAMNP NTVFDAKRLIGRRISD SVQSDMKLWPFKVI GP +KPMI VNYK E+KQF+A
Sbjct: 310 QVAMNPTNTVFDAKRLIGRRISDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQFSA 489
Query: 121 EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 180
EEISSMVL+KM+EIAEAYLG+T+KNAVVTVPAYFNDSQRQATKDAGVI+GLNV+RIINEP
Sbjct: 490 EEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIINEP 669
Query: 181 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 240
TAAAIAYGLDKKATS GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD
Sbjct: 670 TAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 849
Query: 241 NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVDFY 300
NRMVNHFVQEFKRKNKKDISGNPRALRR+RTACERAKRTLSSTAQTTIEIDSLFEG+DFY
Sbjct: 850 NRMVNHFVQEFKRKNKKDISGNPRALRRVRTACERAKRTLSSTAQTTIEIDSLFEGIDFY 1029
Query: 301 TTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQDFF 360
TTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSV DVVLVGGSTRIPKVQQLLQDFF
Sbjct: 1030TTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVDDVVLVGGSTRIPKVQQLLQDFF 1209
Query: 361 NGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLI 420
NGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLI
Sbjct: 1210NGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLI 1389
Query: 421 PRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERARTRDNNLLGKFELSGIPPAPRGVPQI 480
PRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGER RTRDNNLLGKFELSGIPPAPRGVPQI
Sbjct: 1390PRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRGVPQI 1569
Query: 481 TVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVEEAEKYKAEDEEHKK 540
TVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKE+IEKMV+EAEKYK+EDEEHK+
Sbjct: 1570TVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEHKR 1749
Query: 541 KVDAKNALENYAYNMRNTIKDEKIGSKLSPEDKKKIDDAIDAAIQWLDSNQLAEADEFED 600
KV+AKNALENYAYNMRNTIKD+KI SKLS +DKKKI+DAI+ AIQWLD NQL EADEFED
Sbjct: 1750KVEAKNALENYAYNMRNTIKDDKIASKLSADDKKKIEDAIEGAIQWLDGNQLGEADEFED 1929
Query: 601 KMKELESLCNPIIAKMYQGGGAPDMGGGMDDEVPPA--AGGAGPKIEEVD 648
KMKELE +CNPIIA+MYQG G D GG MD++ P A GAGPKIEEVD
Sbjct: 1930KMKELEGICNPIIARMYQGAGG-DAGGAMDEDGPAAGSGSGAGPKIEEVD 2076
>TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protein 4
precursor (BiP 4) (78 kDa glucose-regulated protein
homolog 4) (GRP 78-4)., partial (94%)
Length = 2265
Score = 809 bits (2090), Expect = 0.0
Identities = 407/629 (64%), Positives = 505/629 (79%), Gaps = 3/629 (0%)
Frame = +2
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNP 66
G IGIDLGTTYSCVGV+++ VEIIANDQGNR TPS+V+FTD ERLIG+AAKN A+NP
Sbjct: 155 GTVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAVNP 334
Query: 67 VNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYK-GEDKQFAAEEISS 125
T+FD KRLIGR+ +D VQ DMKL P+K++ G KP I V K GE K F+ EE+S+
Sbjct: 335 ERTIFDVKRLIGRKFADKEVQRDMKLVPYKIVNKDG-KPYIQVRVKDGETKVFSPEEVSA 511
Query: 126 MVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAI 185
M+L KM+E AEA+LG T+++AVVTVPAYFND+QRQATKDAGVIAGLNV RIINEPTAAAI
Sbjct: 512 MILTKMKETAEAFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAAAI 691
Query: 186 AYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVN 245
AYGLDKK GEKN+L+FDLGGGTFDVS+LTI+ G+FEV +T GDTHLGGEDFD R++
Sbjct: 692 AYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLSTNGDTHLGGEDFDQRIME 862
Query: 246 HFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVDFYTTITR 305
+F++ K+K+ KDIS + RAL +LR ERAKR LSS Q +EI+SLF+GVDF +TR
Sbjct: 863 YFIKLIKKKHSKDISKDNRALGKLRRESERAKRALSSQHQVRVEIESLFDGVDFSEPLTR 1042
Query: 306 ARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQDFFNGKEL 365
ARFEELN DLFRK M PV+K + DA + KN + ++VLVGGSTRIPKVQQLL+D+F+GKE
Sbjct: 1043ARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTRIPKVQQLLKDYFDGKEP 1222
Query: 366 CKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTT 425
K +NPDEAVA+GAAVQ +ILSGEG E+ +D+LLLDV PL+LG+ET GGVMT LIPRNT
Sbjct: 1223NKGVNPDEAVAFGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTKLIPRNTV 1402
Query: 426 IPTKKEQVFSTYSDNQPGVLIQVYEGERARTRDNNLLGKFELSGIPPAPRGVPQITVCFD 485
IPTKK QVF+TY D Q V IQV+EGER+ T+D LLG F+LSGIPPAPRG PQI V F+
Sbjct: 1403IPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGNFDLSGIPPAPRGTPQIEVTFE 1582
Query: 486 IDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVEEAEKYKAEDEEHKKKVDAK 545
+DANGILNV AEDK TG+ KITITN+KGRLS+EEI++MV EAE++ ED++ K+++DA+
Sbjct: 1583VDANGILNVRAEDKGTGKSEKITITNEKGRLSQEEIDRMVREAEEFAEEDKKVKERIDAR 1762
Query: 546 NALENYAYNMRNTIKD-EKIGSKLSPEDKKKIDDAIDAAIQWLDSNQLAEADEFEDKMKE 604
NALE Y YNM+N I D +K+ KL ++K+KI+ A+ A++WLD NQ E +EFE+K+KE
Sbjct: 1763NALETYVYNMKNQISDKDKLADKLESDEKEKIEAAVKEALEWLDDNQTVEKEEFEEKLKE 1942
Query: 605 LESLCNPIIAKMYQ-GGGAPDMGGGMDDE 632
+E++CNPII +YQ GGAP G ++
Sbjct: 1943VEAVCNPIITAVYQRSGGAPGGASGEGED 2029
>TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chaperone
HSC71.0 - garden pea, partial (59%)
Length = 1336
Score = 706 bits (1823), Expect = 0.0
Identities = 355/386 (91%), Positives = 368/386 (94%), Gaps = 1/386 (0%)
Frame = +2
Query: 264 RALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVDFYTTITRARFEELNMDLFRKCMEPV 323
RALRRLRTACERAKRTLSSTAQTTIEIDSLFEG+DFY+ ITRARFEELNMDLFRKCMEPV
Sbjct: 2 RALRRLRTACERAKRTLSSTAQTTIEIDSLFEGIDFYSPITRARFEELNMDLFRKCMEPV 181
Query: 324 EKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQA 383
EKCLRDAKMDK SVHDVVLVGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQA
Sbjct: 182 EKCLRDAKMDKKSVHDVVLVGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQA 361
Query: 384 AILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPG 443
AILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPG
Sbjct: 362 AILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPG 541
Query: 444 VLIQVYEGERARTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQ 503
VLIQV+EGER RTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQ
Sbjct: 542 VLIQVFEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQ 721
Query: 504 KNKITITNDKGRLSKEEIEKMVEEAEKYKAEDEEHKKKVDAKNALENYAYNMRNTIKDEK 563
KNKITITNDKGRLSKE+IEKMV+EAEKYK+EDEEHKKKV+AKNALENYAYNMRNTIKDEK
Sbjct: 722 KNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEHKKKVEAKNALENYAYNMRNTIKDEK 901
Query: 564 IGSKLSPEDKKKIDDAIDAAIQWLDSNQLAEADEFEDKMKELESLCNPIIAKMYQGGGAP 623
I KL +DKKKI+D I+AAIQWLD+NQLAEADEFEDKMKELE +CNPIIAKMYQGG P
Sbjct: 902 IAGKLDSDDKKKIEDTIEAAIQWLDANQLAEADEFEDKMKELEGVCNPIIAKMYQGGAGP 1081
Query: 624 DMGGGM-DDEVPPAAGGAGPKIEEVD 648
DMG DD+ P AGGAGPKIEEVD
Sbjct: 1082DMGAAPGDDDAPSHAGGAGPKIEEVD 1159
>TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (53%)
Length = 1189
Score = 591 bits (1523), Expect = e-169
Identities = 300/352 (85%), Positives = 321/352 (90%), Gaps = 6/352 (1%)
Frame = +3
Query: 303 ITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQDFFNG 362
ITRARFEELNMDLFRKCMEPVEKCLRDAK+DK+ VH+VVLVGGSTRIPKVQQLLQDFFNG
Sbjct: 3 ITRARFEELNMDLFRKCMEPVEKCLRDAKIDKSHVHEVVLVGGSTRIPKVQQLLQDFFNG 182
Query: 363 KELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPR 422
KELCKSINPDEAVAYGAAVQAAILSGEG+EKVQDLLLLDVTPLSLGLETAGGVMT LIPR
Sbjct: 183 KELCKSINPDEAVAYGAAVQAAILSGEGDEKVQDLLLLDVTPLSLGLETAGGVMTTLIPR 362
Query: 423 NTTIPTKKEQVFSTYSDNQPGVLIQVYEGERARTRDNNLLGKFELSGIPPAPRGVPQITV 482
NTTIPTKKEQ+FSTYSDNQPGVLIQV+EGERART+DNNLLGKFEL+GIPPAPRGVPQI V
Sbjct: 363 NTTIPTKKEQIFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELTGIPPAPRGVPQINV 542
Query: 483 CFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVEEAEKYKAEDEEHKKKV 542
CFDIDANGILNVSAEDKT G KNKITITNDKGRLSKEEIEKMV++AEKYKAEDEE KKKV
Sbjct: 543 CFDIDANGILNVSAEDKTAGVKNKITITNDKGRLSKEEIEKMVKDAEKYKAEDEEVKKKV 722
Query: 543 DAKNALENYAYNMRNTIKDEKIGSKLSPEDKKKIDDAIDAAIQWLDSNQLAEADEFEDKM 602
+AKN++ENYAYNMRNTIKDEKIG KLS EDK+KI+ A++ AIQWL+ NQ+AE DEFEDK
Sbjct: 723 EAKNSIENYAYNMRNTIKDEKIGGKLSHEDKEKIEKAVEDAIQWLEGNQMAEVDEFEDKQ 902
Query: 603 KELESLCNPIIAKMYQGGGAPD--MGGGMDDEVPPAA----GGAGPKIEEVD 648
KELE +CNPIIAKMYQGG D MG GM A+ GAGPKIEEVD
Sbjct: 903 KELEGICNPIIAKMYQGGAGGDVPMGDGMPGGGGGASNGTGSGAGPKIEEVD 1058
>TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa protein
mitochondrial precursor. [Kidney bean French bean]
{Phaseolus vulgaris}, complete
Length = 2371
Score = 560 bits (1443), Expect = e-160
Identities = 310/638 (48%), Positives = 417/638 (64%), Gaps = 10/638 (1%)
Frame = +2
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDS-ERLIGDAAKNQVAMNPVN 68
IGIDLGTT SCV V + +++ N +G RTTPS VAFT E L+G AK Q NP N
Sbjct: 263 IGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPEN 442
Query: 69 TVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVL 128
T+ AKRLIGRR D Q +MK+ P+K++ P + + + +Q++ +I + VL
Sbjct: 443 TISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWV----EAKGQQYSPSQIGAFVL 610
Query: 129 IKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYG 188
KM+E AEAYLG TV AV+TVPAYFND+QRQATKDAG IAGL VLRIINEPTAAA++YG
Sbjct: 611 TKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYG 790
Query: 189 LDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 248
++K+ + +FDLGGGTFDVS+L I G+FEVKAT GDT LGGEDFDN +++ V
Sbjct: 791 MNKEGL------IAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLV 952
Query: 249 QEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVD----FYTTIT 304
EFKR D+S + AL+RLR A E+AK LSST+QT I + + T+T
Sbjct: 953 NEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLT 1132
Query: 305 RARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQDFFNGKE 364
R++FE L +L + P + CL+DA + + +V+LVGG TR+PKVQ+++ F GK
Sbjct: 1133RSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIF-GKS 1309
Query: 365 LCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNT 424
CK +NPDEAVA GAA+Q IL G+ V++LLLLDVTPLSLG+ET GG+ T LI RNT
Sbjct: 1310PCKGVNPDEAVAMGAALQGGILRGD----VKELLLLDVTPLSLGIETLGGIFTRLINRNT 1477
Query: 425 TIPTKKEQVFSTYSDNQPGVLIQVYEGERARTRDNNLLGKFELSGIPPAPRGVPQITVCF 484
TIPTKK QVFST +DNQ V I+V +GER DN +LG+FEL G+PPAPRG+PQI V F
Sbjct: 1478TIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTF 1657
Query: 485 DIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVEEAEKYKAEDEEHKKKVDA 544
DIDANGI+ VSA+DK+TG++ +ITI G LS +EI+ MV+EAE + +D+E K +D
Sbjct: 1658DIDANGIVTVSAKDKSTGKEQQITI-KSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDI 1834
Query: 545 KNALENYAYNMRNTIKDEKIGSKLSPEDKKKIDDAIDAAIQWLDSNQLAEADEFEDKMKE 604
+N+ + Y++ ++ + + K+ E K+I+DA+ + A A + D++K
Sbjct: 1835RNSADTSIYSIEKSLSEYR--EKIPAEVAKEIEDAV-------SDLRSAMAGDSADEIKS 1987
Query: 605 LESLCNPIIAKM--YQGGGA---PDMGGGMDDEVPPAA 637
N ++K+ + GG+ P G + P A
Sbjct: 1988KLDAANKAVSKIGEHMSGGSSSGPSSDGSQGGDQAPEA 2101
>TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chaperone PHSP1
precursor mitochondrial - garden pea, partial (98%)
Length = 2422
Score = 546 bits (1407), Expect = e-156
Identities = 311/647 (48%), Positives = 411/647 (63%), Gaps = 11/647 (1%)
Frame = +1
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDS-ERLIGDAAKNQVAMNPVN 68
IGIDLGTT SCV + + ++I N +G RTTPS VAF E L+G AK Q NP N
Sbjct: 247 IGIDLGTTNSCVSLMEGKNPKVIENSEGARTTPSVVAFNQKGELLVGTPAKRQAVTNPTN 426
Query: 69 TVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVL 128
T+F KRLIGRR D Q +MK+ P+K++ P + +N +Q++ +I + VL
Sbjct: 427 TLFGTKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEIN----KQQYSPSQIGAFVL 594
Query: 129 IKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYG 188
KM+E AEAYLG T+ AVVTVPAYFND+QRQATKDAG IAGL V RIINEPTAAA++YG
Sbjct: 595 TKMKETAEAYLGKTISKAVVTVPAYFNDAQRQATKDAGRIAGLEVKRIINEPTAAALSYG 774
Query: 189 LDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 248
++ K E + +FDLGGGTFDVS+L I G+FEVKAT GDT LGGEDFDN +++ V
Sbjct: 775 MNNK-----EGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLV 939
Query: 249 QEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVD----FYTTIT 304
EFKR + D++ + AL+RLR A E+AK LSST+QT I + + T+T
Sbjct: 940 SEFKRTDSIDLAKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLT 1119
Query: 305 RARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQDFFNGKE 364
R++FE L +L + P + CL+DA + V +V+LVGG TR+PKVQ+++ + F GK
Sbjct: 1120RSKFEALVNNLIERTKAPCKSCLKDANISIKDVDEVLLVGGMTRVPKVQEVVSEIF-GKS 1296
Query: 365 LCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNT 424
K +NPDEAVA GAA+Q IL G+ V++LLLLDVTPLSLG+ET GG+ T LI RNT
Sbjct: 1297PSKGVNPDEAVAMGAALQGGILRGD----VKELLLLDVTPLSLGIETLGGIFTRLISRNT 1464
Query: 425 TIPTKKEQVFSTYSDNQP--GVLIQVYEGERARTRDNNLLGKFELSGIPPAPRGVPQITV 482
TIPTKK QVFST +DNQ G EG R DN LG+F+L GIPPAPRG+PQI V
Sbjct: 1465TIPTKKSQVFSTAADNQTQRGYXRCSQEGXREMAADNKSLGEFDLVGIPPAPRGLPQIEV 1644
Query: 483 CFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVEEAEKYKAEDEEHKKKV 542
FDIDANGI+ VSA+DK+TG++ +ITI G LS +EI MV+EAE + D+E K +
Sbjct: 1645TFDIDANGIVTVSAKDKSTGKEQQITI-RSSGGLSDDEINNMVKEAELHAQRDQERKALI 1821
Query: 543 DAKNALENYAYNMRNTIKD--EKIGSKLSPEDKKKIDDAIDAAIQWLDSNQLAEADEFED 600
D KN+ + Y++ ++ + EKI S+++ E + + D + A E D
Sbjct: 1822DIKNSADTSIYSIEKSLSEYREKIPSEVAKEIENSVSDL-----------RTAMEGESVD 1968
Query: 601 KMKELESLCNPIIAKM--YQGGGAPDMGGGMDDEVPPAAGGAGPKIE 645
++K N ++K+ + GG+ GG D P G P+ E
Sbjct: 1969EIKTKLDAANKAVSKIGQHMSGGS---SGGSSDGGSPGGGDQAPEAE 2100
>TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa protein
(HSP70). {Neurospora crassa}, partial (54%)
Length = 1258
Score = 525 bits (1352), Expect(2) = e-155
Identities = 267/333 (80%), Positives = 299/333 (89%)
Frame = +2
Query: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPVN 68
AIGIDLGTTYSCVG + +D++EIIANDQGNRTTPSYVAF D+ERLIGDAAKNQVAMNP N
Sbjct: 188 AIGIDLGTTYSCVGRYANDKIEIIANDQGNRTTPSYVAFNDTERLIGDAAKNQVAMNPHN 367
Query: 69 TVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVL 128
TVFDAKRLIGR+ SD+ VQ+DMK +PFKVI G KP I V +KGE+K F EEIS+MVL
Sbjct: 368 TVFDAKRLIGRKFSDSEVQADMKHFPFKVI-DKGGKPNIEVEFKGENKTFTPEEISAMVL 544
Query: 129 IKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYG 188
+KMRE AEAYLG V NAV+TVPAYFNDSQRQATKDAG+IAGLNVLRIINEPTAAAIAYG
Sbjct: 545 VKMRETAEAYLGGQVTNAVITVPAYFNDSQRQATKDAGLIAGLNVLRIINEPTAAAIAYG 724
Query: 189 LDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 248
LDKKA GE+NVLIFDLGGGTFDVSLLTIEEGIFEVK+TAGDTHLGGEDFDNR+VNHFV
Sbjct: 725 LDKKAE--GERNVLIFDLGGGTFDVSLLTIEEGIFEVKSTAGDTHLGGEDFDNRLVNHFV 898
Query: 249 QEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVDFYTTITRARF 308
EFKRK+KKD+S N RALRRLRTACERAKRTLSS+AQT+IEIDSLFEG+DFYT+ITRARF
Sbjct: 899 NEFKRKHKKDLSSNARALRRLRTACERAKRTLSSSAQTSIEIDSLFEGIDFYTSITRARF 1078
Query: 309 EELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVV 341
EEL DLFR ++PV++ L DAK+DK+ VH++V
Sbjct: 1079EELCQDLFRSTIQPVDRVLSDAKIDKSQVHEIV 1177
Score = 41.2 bits (95), Expect(2) = e-155
Identities = 16/22 (72%), Positives = 21/22 (94%)
Frame = +3
Query: 343 VGGSTRIPKVQQLLQDFFNGKE 364
VGGSTRIP++Q+L+ D+FNGKE
Sbjct: 1182 VGGSTRIPRIQKLISDYFNGKE 1247
>TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (44%)
Length = 955
Score = 507 bits (1305), Expect = e-144
Identities = 255/282 (90%), Positives = 267/282 (94%)
Frame = +2
Query: 6 EGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMN 65
EG AIGIDLGTTYSCVGVWQ+DRVEII NDQGNRTTPSYVAFTD+ERLIGDAAKNQVAMN
Sbjct: 110 EGKAIGIDLGTTYSCVGVWQNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAKNQVAMN 289
Query: 66 PVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISS 125
P NTVFDAKRLIGRR SD SVQ+DMKLWPFKV+ GP EKPMI VNYKGE+K+FAAEEISS
Sbjct: 290 PQNTVFDAKRLIGRRFSDESVQNDMKLWPFKVVPGPAEKPMIVVNYKGEEKKFAAEEISS 469
Query: 126 MVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAI 185
MVLIKMRE+AEA+LG VKNAVVTVPAYFNDSQRQATKDAG I+GLNVLRIINEPTAAAI
Sbjct: 470 MVLIKMREVAEAFLGHPVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEPTAAAI 649
Query: 186 AYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVN 245
AYGLDKKA+ GE+NVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVN
Sbjct: 650 AYGLDKKASRKGEQNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVN 829
Query: 246 HFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTT 287
HFV EF+RKNKKDISGN RALRRLRTACERAKRTLSSTAQTT
Sbjct: 830 HFVSEFRRKNKKDISGNARALRRLRTACERAKRTLSSTAQTT 955
>TC85395 homologue to GP|6911553|emb|CAB72130.1 heat shock protein 70
{Cucumis sativus}, partial (35%)
Length = 723
Score = 448 bits (1152), Expect = e-126
Identities = 225/226 (99%), Positives = 226/226 (99%)
Frame = +3
Query: 395 QDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERA 454
QDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERA
Sbjct: 3 QDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERA 182
Query: 455 RTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKG 514
RTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKG
Sbjct: 183 RTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKG 362
Query: 515 RLSKEEIEKMVEEAEKYKAEDEEHKKKVDAKNALENYAYNMRNTIKDEKIGSKLSPEDKK 574
RLSKEEIEKMVEEAEKYKAEDEEHKKKVDAKNALENYAYNMRNTIKDEKIGSKLSPEDKK
Sbjct: 363 RLSKEEIEKMVEEAEKYKAEDEEHKKKVDAKNALENYAYNMRNTIKDEKIGSKLSPEDKK 542
Query: 575 KIDDAIDAAIQWLDSNQLAEADEFEDKMKELESLCNPIIAKMYQGG 620
KIDDAIDAAIQWLDSNQLAEADEF+DKMKELESLCNPIIAKMYQGG
Sbjct: 543 KIDDAIDAAIQWLDSNQLAEADEFQDKMKELESLCNPIIAKMYQGG 680
Score = 34.3 bits (77), Expect = 0.15
Identities = 14/33 (42%), Positives = 21/33 (63%)
Frame = +1
Query: 598 FEDKMKELESLCNPIIAKMYQGGGAPDMGGGMD 630
F+ + + L + + + +GGGAPDMGGGMD
Sbjct: 613 FKIR*RNLRAFATQSLPRCIKGGGAPDMGGGMD 711
>TC85391 homologue to PIR|T06598|T06598 dnaK-type molecular chaperone BiP-A
- soybean, partial (52%)
Length = 1231
Score = 434 bits (1117), Expect = e-122
Identities = 213/340 (62%), Positives = 271/340 (79%), Gaps = 2/340 (0%)
Frame = +1
Query: 295 EGVDFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQ 354
+GV F +TRARFEELN DLFRK M PV+K + DA + KN + ++VLVGGSTRIPKVQQ
Sbjct: 1 DGVAFSEPLTRARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTRIPKVQQ 180
Query: 355 LLQDFFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGG 414
LL+D+F+GKE K +NPDEAVA+GAAVQ +ILSGEG + +D+LLLDV PL+LG+ET GG
Sbjct: 181 LLKDYFDGKEPNKGVNPDEAVAFGAAVQGSILSGEGGAETKDILLLDVAPLTLGIETVGG 360
Query: 415 VMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERARTRDNNLLGKFELSGIPPAP 474
VMT LIPRNT IPTKK QVF+TY D Q V IQV+EGER+ T+D LLGKF+LSGIPPAP
Sbjct: 361 VMTKLIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLSGIPPAP 540
Query: 475 RGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVEEAEKYKAE 534
RG PQI V F++DANGILNV AEDK TG+ KITITN+KGRLS+EEIE+MV EAE++ E
Sbjct: 541 RGTPQIEVTFEVDANGILNVKAEDKGTGKSEKITITNEKGRLSQEEIERMVREAEEFAEE 720
Query: 535 DEEHKKKVDAKNALENYAYNMRNTIKD-EKIGSKLSPEDKKKIDDAIDAAIQWLDSNQLA 593
D++ K+++DA+NALE Y YNM+N + D +K+ KL ++K+KI+ A+ A++WLD NQ
Sbjct: 721 DKKVKERIDARNALETYVYNMKNQVSDKDKLADKLESDEKEKIETAVKEALEWLDDNQSV 900
Query: 594 EADEFEDKMKELESLCNPIIAKMYQ-GGGAPDMGGGMDDE 632
E +E+E+K+KE+E++CNPII +YQ GGAP G +DE
Sbjct: 901 EKEEYEEKLKEVEAVCNPIITAVYQRSGGAPGGGASGEDE 1020
>BQ139103 homologue to GP|2655420|gb| heat shock cognate protein HSC70
{Brassica napus}, partial (29%)
Length = 628
Score = 366 bits (939), Expect = e-101
Identities = 180/191 (94%), Positives = 188/191 (98%)
Frame = +2
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN
Sbjct: 56 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 235
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
QVAMNP+NTVFDAKRLIGRR SDASVQSDMKLWPFK+I+GP EKP+IGVNYKGEDK+FAA
Sbjct: 236 QVAMNPINTVFDAKRLIGRRFSDASVQSDMKLWPFKIISGPAEKPLIGVNYKGEDKEFAA 415
Query: 121 EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 180
EEISSMVL+KMREIAEAYLGS +KNAVVTVPAYFNDSQRQATKDAGVIAGLNV+RIINEP
Sbjct: 416 EEISSMVLMKMREIAEAYLGSAIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEP 595
Query: 181 TAAAIAYGLDK 191
TAAAIAYGLDK
Sbjct: 596 TAAAIAYGLDK 628
>BQ752418 homologue to SP|P78695|GR78 78 kDa glucose-regulated protein
homolog precursor (GRP 78), partial (47%)
Length = 1032
Score = 336 bits (862), Expect(3) = e-101
Identities = 167/288 (57%), Positives = 224/288 (76%), Gaps = 2/288 (0%)
Frame = -1
Query: 349 IPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLG 408
IPKVQ L++++F GK+ K INPDEAVA+GAAVQA +LSGE E ++++L+DV PL+LG
Sbjct: 900 IPKVQSLIEEYFGGKKASKGINPDEAVAFGAAVQAGVLSGE--EGTEEIVLMDVNPLTLG 727
Query: 409 LETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERARTRDNNLLGKFELS 468
+ET GGVMT LI RNT IPT+K Q+FST +DNQP VLIQV+EGER+ T+DNN LGKFEL+
Sbjct: 726 IETTGGVMTKLITRNTPIPTRKSQIFSTAADNQPVVLIQVFEGERSLTKDNNQLGKFELT 547
Query: 469 GIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVEEA 528
GIPPAPRGVPQI V F++DANGIL VSA DK TG++ ITITNDKGRL+KEEI++MVEEA
Sbjct: 546 GIPPAPRGVPQIEVSFELDANGILKVSAHDKGTGKQESITITNDKGRLTKEEIDRMVEEA 367
Query: 529 EKYKAEDEEHKKKVDAKNALENYAYNMRNTIKDEK-IGSKLSPEDKKKIDDAIDAAIQWL 587
EKY ED+ +++++A+N LENYA++++N + DE+ +G K+ EDK+ I +A+ WL
Sbjct: 366 EKYAEEDKATRERIEARNGLENYAFSLKNQVNDEEGLGGKIDDEDKETILEAVKETNDWL 187
Query: 588 DSN-QLAEADEFEDKMKELESLCNPIIAKMYQGGGAPDMGGGMDDEVP 634
+ N A ++FE++ ++L ++ PI +KMYQG G G G +DE P
Sbjct: 186 EENGATANTEDFEEQKEKLSNVAYPITSKMYQGAG----GAGGEDEPP 55
Score = 44.7 bits (104), Expect(3) = e-101
Identities = 20/35 (57%), Positives = 26/35 (74%)
Frame = -3
Query: 317 RKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPK 351
R+ PVE+ L+DAK+ K V D+VLVGGSTR P+
Sbjct: 997 RRLSSPVEQVLKDAKVKKEDVDDIVLVGGSTRYPQ 893
Score = 28.9 bits (63), Expect(3) = e-101
Identities = 11/17 (64%), Positives = 15/17 (87%)
Frame = -2
Query: 306 ARFEELNMDLFRKCMEP 322
A+FEELNMDL +K ++P
Sbjct: 1031 AKFEELNMDLLKKTLKP 981
>TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic reticulum
HSC70-cognate binding protein precursor {Glycine max},
partial (38%)
Length = 962
Score = 313 bits (801), Expect = 2e-85
Identities = 161/228 (70%), Positives = 187/228 (81%), Gaps = 1/228 (0%)
Frame = +1
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNP 66
G IGIDLGTTYSCVGV+++ VEIIANDQGNR TPS+V+FTD ERLIG+AAKN A+NP
Sbjct: 289 GTVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAVNP 468
Query: 67 VNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYK-GEDKQFAAEEISS 125
T+FD KRLIGR+ D VQ DMKL P+K++ G KP I V K GE K F+ EEIS+
Sbjct: 469 ERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNRDG-KPYIQVRVKDGETKVFSPEEISA 645
Query: 126 MVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAI 185
M+L KM+E AE +LG T+++AVVTVPAYFND+QRQATKDAGVIAGLNV RIINEPTAAAI
Sbjct: 646 MILGKMKETAEGFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAAAI 825
Query: 186 AYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTH 233
AYGLDKK GEKN+L+FDLGGGTFDVS+LTI+ G+FEV AT GDTH
Sbjct: 826 AYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTH 960
>TC89238 similar to SP|Q01877|HS71_PUCGR Heat shock protein HSS1. {Puccinia
graminis}, partial (32%)
Length = 1015
Score = 300 bits (767), Expect = 2e-81
Identities = 152/224 (67%), Positives = 176/224 (77%), Gaps = 9/224 (4%)
Frame = +1
Query: 434 FSTYSDNQPGVLIQVYEGERARTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILN 493
FSTYSDNQPGVLIQVYEGER RTRDNNLLGKFEL+GIPPAPRGVPQI V FDIDANGILN
Sbjct: 1 FSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELTGIPPAPRGVPQIEVTFDIDANGILN 180
Query: 494 VSAEDKTTGQKNKITITNDKGRLSKEEIEKMVEEAEKYKAEDEEHKKKVDAKNALENYAY 553
VSA DKTTG+ NKITITNDKGRLSKEEIE+MV EAEKYKA+DE+ +++ ++N LE+YAY
Sbjct: 181 VSAVDKTTGRSNKITITNDKGRLSKEEIERMVAEAEKYKADDEKVAQRIQSRNGLESYAY 360
Query: 554 NMRNTIKDEKIGSKLSPEDKKKIDDAIDAAIQWLDSNQLAEADEFEDKMKELESLCNPII 613
N+RNT++DEKI KL P DKKK++DAI +I WLD+NQ AE +E++ K K LE + NPI+
Sbjct: 361 NLRNTLQDEKISGKLDPADKKKLEDAIQESITWLDNNQEAEKEEYDHKQKSLEEIANPIM 540
Query: 614 AKMYQG-GGAPDMGGGMD--------DEVPPAAGGAGPKIEEVD 648
K+Y G GGAP GG PA AGP IEEVD
Sbjct: 541 MKLYGGAGGAPPGAGGFPPGGFPGGAPGGAPADDNAGPTIEEVD 672
>TC80997 similar to PIR|B84729|B84729 70kD heat shock protein [imported] -
Arabidopsis thaliana, partial (91%)
Length = 1717
Score = 181 bits (459), Expect(2) = 6e-76
Identities = 93/232 (40%), Positives = 143/232 (61%), Gaps = 1/232 (0%)
Frame = +1
Query: 264 RALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVDFYTTITRARFEELNMDLFRKCMEPV 323
+++ LR A + A LS+ + +++D L + + RA FEE+N ++F KC +
Sbjct: 772 KSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKICKVVDRAEFEEVNKEVFEKCESLI 948
Query: 324 EKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQA 383
+CL DAK++ ++DV+LVGG + IP+V+ L+ + E+ K INP E GA ++
Sbjct: 949 IQCLHDAKVEVGDINDVILVGGCSYIPRVENLVTNLCKITEVYKGINPLEGAVCGATMEG 1128
Query: 384 AILSGEGNEKVQ-DLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQP 442
A+ SG + DLL + TPL++G+ G +IPRNTT+P +K+ +F+T DNQ
Sbjct: 1129 AVASGISDPFGNLDLLTIQATPLAIGIRADGNKFVPVIPRNTTMPARKDLLFTTIHDNQT 1308
Query: 443 GVLIQVYEGERARTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNV 494
LI VYEGE + +N+LLG F++ GIP AP+GVP+I+VC DIDA +L V
Sbjct: 1309 EALILVYEGEGKKAEENHLLGYFKIMGIPAAPKGVPEISVCMDIDAANVLRV 1464
Score = 122 bits (305), Expect(2) = 6e-76
Identities = 84/252 (33%), Positives = 131/252 (51%), Gaps = 13/252 (5%)
Frame = +3
Query: 15 GTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNPVNTVFD 72
GT+ V VW VE++ N + + S+V F D G ++ ++ M +T+F+
Sbjct: 3 GTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLFGDTIFN 182
Query: 73 AKRLIGRRISDASVQSDMKLWPFKV-IAGPGEKPMIGVNYKGEDKQFAAEEISSMVLIKM 131
KRLIGR +D V + L PF V G +P I + EE+ +M L+++
Sbjct: 183 MKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLAMFLVEL 359
Query: 132 REIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYGLDK 191
R + E +L ++N V+TVP F+ Q + A +AGL+VLR++ EPTA A+ YG +
Sbjct: 360 RLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVALLYGQQQ 539
Query: 192 KATSV------GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVN 245
+ S EK LIF++G G DV++ G+ ++KA AG T +GGED M+
Sbjct: 540 QKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDLLQNMMR 716
Query: 246 HFVQE----FKR 253
H + + FKR
Sbjct: 717 HLLPDSENIFKR 752
>TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chaperone hsc70-3
- tomato, partial (27%)
Length = 667
Score = 276 bits (705), Expect = 2e-74
Identities = 137/192 (71%), Positives = 160/192 (82%)
Frame = +3
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
M+G+G+ AIGIDLGTTYSCV V ++ ++II ND G RTTPS+VAF DSER+IGDAA N
Sbjct: 81 MSGRGKKVAIGIDLGTTYSCVAVCKNGEIDIIVNDLGKRTTPSFVAFKDSERMIGDAAFN 260
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
A NP NT+FDAKRLIGR+ SD VQSD+KLWPFKVI +KPMI VNY E+K FAA
Sbjct: 261 IAASNPTNTIFDAKRLIGRKFSDPIVQSDVKLWPFKVIGDLNDKPMIVVNYNDEEKHFAA 440
Query: 121 EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 180
EEISSMVL+KMREIAE +LGS V++ V+TVPAYFNDSQRQ+T+DAG IAGLNV+RIINEP
Sbjct: 441 EEISSMVLVKMREIAETFLGSIVEDVVITVPAYFNDSQRQSTRDAGAIAGLNVMRIINEP 620
Query: 181 TAAAIAYGLDKK 192
TAAAIAYG + K
Sbjct: 621 TAAAIAYGFNTK 656
>TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock protein
{Arabidopsis thaliana}, partial (92%)
Length = 3027
Score = 270 bits (691), Expect = 1e-72
Identities = 152/438 (34%), Positives = 238/438 (53%), Gaps = 13/438 (2%)
Frame = +1
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPVNT 69
+G D G V V + ++++ ND+ R TP+ V F D +R IG A MNP N+
Sbjct: 247 VGFDFGNESCIVAVARQRGIDVVLNDESKRETPAIVCFGDKQRFIGTAGAASTMMNPKNS 426
Query: 70 VFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVLI 129
+ KRLIG++ +D +Q D+K PF V GP P+I Y GE ++F A ++ M+L
Sbjct: 427 ISQIKRLIGKKFADPELQRDLKSLPFNVTEGPDGYPLIHARYLGESREFTATQVFGMMLS 606
Query: 130 KMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYGL 189
++EIA+ L + V + + +P YF D QR++ DA IAGL+ L +I+E TA A+AYG+
Sbjct: 607 NLKEIAQKNLNAAVVDCCIGIPVYFTDLQRRSVLDAATIAGLHPLHLIHETTATALAYGI 786
Query: 190 DKKATSVGE-KNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 248
K E NV D+G + V + ++G V + + D LGG DFD + +HF
Sbjct: 787 YKTDLPENEWLNVAFVDVGHASMQVCIAGFKKGQLHVLSHSYDRSLGGRDFDEALFHHFA 966
Query: 249 QEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVDFYTTITRARF 308
+FK + K D+ N RA RLR ACE+ K+ LS+ + + I+ L + D I R F
Sbjct: 967 AKFKEEYKIDVYQNARACLRLRAACEKLKKVLSANPEAPLNIECLMDEKDVRGFIKRDDF 1146
Query: 309 EELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQDFFNGKELCKS 368
E+L++ + + P+EK L +A + ++H V +VG +R+P + ++L +FF KE ++
Sbjct: 1147EQLSLPILERVKGPLEKALAEAGLTVENIHMVEVVGSGSRVPAINKILTEFFK-KEPRRT 1323
Query: 369 INPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGL------------ETAGGVM 416
+N E VA GAA+Q AILS KV++ + + P S+ L E+
Sbjct: 1324MNASECVARGAALQCAILS--PTFKVREFQVNESFPFSVSLSWKYSGSDAPDSESDNKQS 1497
Query: 417 TVLIPRNTTIPTKKEQVF 434
T++ P+ IP+ K F
Sbjct: 1498TIVFPKGNPIPSSKVLTF 1551
Score = 30.8 bits (68), Expect = 1.7
Identities = 14/40 (35%), Positives = 22/40 (55%)
Frame = +3
Query: 523 KMVEEAEKYKAEDEEHKKKVDAKNALENYAYNMRNTIKDE 562
K E + +D ++ D KNA+E Y Y+MRN + D+
Sbjct: 2040 KGCREGVEMALQDRVMEETKDNKNAIEAYVYDMRNKLNDK 2159
>TC88922 homologue to SP|Q02028|HS7S_PEA Stromal 70 kDa heat shock-related
protein chloroplast precursor. [Garden pea] {Pisum
sativum}, partial (42%)
Length = 1092
Score = 221 bits (564), Expect = 5e-58
Identities = 127/284 (44%), Positives = 171/284 (59%), Gaps = 4/284 (1%)
Frame = +2
Query: 369 INPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPT 428
+NPDE VA GAAVQA +L+G+ V D++LLDV+PLSLGLET GGVMT +IPRNTT+PT
Sbjct: 50 VNPDEVVALGAAVQAGVLAGD----VSDIVLLDVSPLSLGLETLGGVMTKIIPRNTTLPT 217
Query: 429 KKEQVFSTYSDNQPGVLIQVYEGERARTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDA 488
K +VFST +D Q V I V +GER RDN LG F L GIPPAPRGVPQI V FDIDA
Sbjct: 218 SKSEVFSTAADGQTSVEINVLQGEREFVRDNKSLGSFRLDGIPPAPRGVPQIEVKFDIDA 397
Query: 489 NGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVEEAEKYKAEDEEHKKKVDAKNAL 548
NGIL+V+A DK TG+K ITIT L +E+E+MV EAE++ ED+E + +D KN
Sbjct: 398 NGILSVAAIDKGTGKKQDITITG-ASTLPGDEVERMVNEAERFSKEDKEKRDAIDTKNQA 574
Query: 549 ENYAYNMRNTIKDEKIGSKLSPEDKKKIDDAIDAAIQWLDSNQLAEADEFEDKMKELESL 608
++ Y +K+ +G K+ K+K++ + ++ D+ +D M L
Sbjct: 575 DSVVYQTEKQLKE--LGEKVPGPVKEKVEAKL---VELKDAISGGSTQTMKDAMAALNQE 739
Query: 609 CNPIIAKMYQGGGAPDMGG----GMDDEVPPAAGGAGPKIEEVD 648
+ +Y GA D G G + ++ G G + +D
Sbjct: 740 VMQLGQSLYNQPGAADAAGPTPPGSESGPTDSSSGKGADGDVID 871
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.134 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,194,710
Number of Sequences: 36976
Number of extensions: 175999
Number of successful extensions: 875
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 793
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 820
length of query: 648
length of database: 9,014,727
effective HSP length: 102
effective length of query: 546
effective length of database: 5,243,175
effective search space: 2862773550
effective search space used: 2862773550
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC147536.1


