

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147517.7 - phase: 0
(485 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
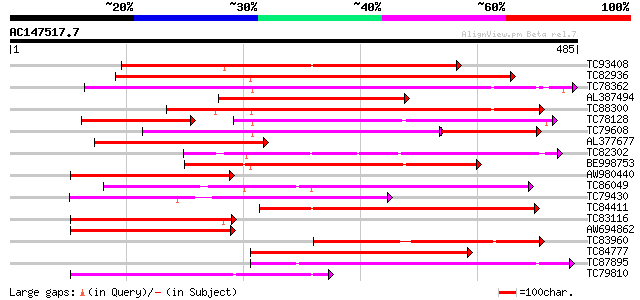
Score E
Sequences producing significant alignments: (bits) Value
TC93408 similar to GP|11933407|dbj|BAB19758. putative nitrate tr... 374 e-104
TC82936 similar to PIR|G84829|G84829 probable PTR2 family peptid... 330 1e-90
TC78362 similar to GP|15391731|emb|CAA93316. nitrite transporter... 293 1e-79
AL387494 weakly similar to GP|17381202|gb putative peptide trans... 260 9e-70
TC88300 similar to GP|9581817|emb|CAC00544.1 putative low-affini... 249 2e-66
TC78128 weakly similar to GP|21928025|gb|AAM78041.1 At1g69870/T1... 179 3e-63
TC79608 similar to GP|20147231|gb|AAM10330.1 At1g68570/F24J5_7 {... 174 1e-60
AL377677 weakly similar to GP|10716608|gb putative peptide trans... 218 3e-57
TC82302 similar to PIR|F84663|F84663 probable nitrate transporte... 213 1e-55
BE998753 similar to PIR|T45958|T45 oligopeptide transporter-like... 210 9e-55
AW980440 similar to GP|17381202|gb putative peptide transporter ... 201 4e-52
TC86049 weakly similar to GP|20466248|gb|AAM20441.1 putative tra... 196 2e-50
TC79430 similar to PIR|F86358|F86358 Similar to peptide transpor... 195 3e-50
TC84411 similar to GP|4102839|gb|AAD01600.1| LeOPT1 {Lycopersico... 187 6e-48
TC83116 similar to GP|11933407|dbj|BAB19758. putative nitrate tr... 178 4e-45
AW694862 similar to GP|10716600|gb putative peptide transport pr... 165 3e-41
TC83960 weakly similar to GP|13605523|gb|AAK32755.1 AT3g53960/F5... 158 4e-39
TC84777 weakly similar to GP|20147231|gb|AAM10330.1 At1g68570/F2... 150 8e-37
TC87895 similar to GP|20466248|gb|AAM20441.1 putative transport ... 142 2e-34
TC79810 similar to GP|11994403|dbj|BAB02362. nitrate transporter... 142 4e-34
>TC93408 similar to GP|11933407|dbj|BAB19758. putative nitrate transporter
NRT1-3 {Glycine max}, partial (44%)
Length = 909
Score = 374 bits (960), Expect = e-104
Identities = 178/293 (60%), Positives = 239/293 (80%), Gaps = 2/293 (0%)
Frame = +1
Query: 96 AQTILVYIQDNVGFALGYGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAM 155
A T+LVYIQDNVG+ LGY +PT+ L +SI++F+ GTP YRH+LP+GS TRM +V VA++
Sbjct: 1 ANTVLVYIQDNVGWTLGYALPTLGLAISIMIFLGGTPFYRHKLPAGSTFTRMARVIVASL 180
Query: 156 RKWKLNVPIDSKELHEVSIEEYTSKGR--YRINHSSSLRFLDKAAVKTGQTSPWMLCTVT 213
RKWK+ VP D+K+L+E+ +EEY KG YRI+ + +LRFLDKA+VKTG TSPWMLCTVT
Sbjct: 181 RKWKVPVPDDTKKLYELDMEEYAKKGSNTYRIDSTPTLRFLDKASVKTGSTSPWMLCTVT 360
Query: 214 QIEETKQMTKMLPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNI 273
+EETKQM +M+P+L+ T +PST+++Q TLF++QGTTLDR +G+ F+IPPA L AFV +
Sbjct: 361 HVEETKQMLRMIPILVATFVPSTMMAQVNTLFVKQGTTLDRHIGS-FKIPPASLAAFVTL 537
Query: 274 FMLISVVIYDRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARE 333
+L+ VV+YDR FV +++R TKNPRGIT+LQR+GIGLV+H I+M+VA + E RL VA+E
Sbjct: 538 SLLVCVVLYDRFFVRIMQRLTKNPRGITLLQRMGIGLVLHTIIMVVASVTENYRLRVAKE 717
Query: 334 NNLLGPLDTIPLTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTS 386
+ L+ +PL+IFIL+PQF LMG AD F+++AK+EFFYDQAP SMKS+GTS
Sbjct: 718 HGLVESGGQVPLSIFILLPQFILMGTADAFLEVAKIEFFYDQAPXSMKSIGTS 876
>TC82936 similar to PIR|G84829|G84829 probable PTR2 family peptide
transporter [imported] - Arabidopsis thaliana, partial
(53%)
Length = 1112
Score = 330 bits (845), Expect = 1e-90
Identities = 163/346 (47%), Positives = 243/346 (70%), Gaps = 4/346 (1%)
Frame = +3
Query: 91 IGTISAQTILVYIQDNVGFALGYGIPTILLVVSILVFVLGTPLYRHRL-PSGSPLTRMVQ 149
+G + A LVYIQ+N+G+ LGYGIPT L++S+++F +GTP+YRH++ S SP +++
Sbjct: 75 LGALIATLGLVYIQENLGWGLGYGIPTAGLILSLVIFYIGTPIYRHKVRTSKSPAKDIIR 254
Query: 150 VFVAAMRKWKLNVPIDSKELHEVSIEEYTSKGRYRINHSSSLRFLDKAAVKTGQT---SP 206
VF+ A + KL +P + ELHE +E +G+ ++ H+ +LRFLDKAA+K T S
Sbjct: 255 VFIVAFKSRKLQLPSNPSELHEFQMEHCVIRGKRQVYHTPTLRFLDKAAIKEDPTTGSSR 434
Query: 207 WMLCTVTQIEETKQMTKMLPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPAC 266
+ TV Q+E K + ML + + T IPSTI +Q TLF++QGTTLDR +G F+IP A
Sbjct: 435 RVPMTVNQVEGAKLILGMLLIWLVTLIPSTIWAQINTLFVKQGTTLDRNLGPDFKIPAAS 614
Query: 267 LIAFVNIFMLISVVIYDRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERK 326
L +FV + ML+SV +YDR+FVP +R+ T +PRGIT+LQRLGIG + +I + +A +E +
Sbjct: 615 LGSFVTLSMLLSVPMYDRLFVPFMRQKTGHPRGITLLQRLGIGFSIQIIAIAIAYAVEVR 794
Query: 327 RLSVARENNLLGPLDTIPLTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTS 386
R+ V +EN++ GP D +P++IF L+PQ+ L+GIAD F I LEFFYDQ+PE M+SLGT+
Sbjct: 795 RMHVIKENHIFGPKDIVPMSIFWLLPQYVLIGIADVFNAIGLLEFFYDQSPEDMQSLGTT 974
Query: 387 YATTTLSIGNFLSTFLLSIVADLTSKNGHKGWILNNLNVSRIDYYY 432
+ T+ + +GNFL++FL+++ +T + K WI +NLN S + YYY
Sbjct: 975 FFTSGIGVGNFLNSFLVTMTDKITGRGDRKSWIADNLNDSHLXYYY 1112
Score = 33.5 bits (75), Expect = 0.19
Identities = 16/63 (25%), Positives = 32/63 (50%), Gaps = 2/63 (3%)
Frame = +2
Query: 67 FDEFEXKERSQKLSFYNWWVFYILIGTISA--QTILVYIQDNVGFALGYGIPTILLVVSI 124
FD+F E+ K SF+NWW+F + ++ +T + + +G + Y +V+S
Sbjct: 2 FDDFNPHEKELKASFFNWWMFTFIFRCLNCNFRTCIYSREFRMGTWIWYSYCWFNIVLSY 181
Query: 125 LVF 127
++
Sbjct: 182 FLY 190
>TC78362 similar to GP|15391731|emb|CAA93316. nitrite transporter {Cucumis
sativus}, partial (88%)
Length = 1871
Score = 293 bits (750), Expect = 1e-79
Identities = 169/435 (38%), Positives = 263/435 (59%), Gaps = 14/435 (3%)
Frame = +3
Query: 65 DQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALGYGIPTILLVVSI 124
DQFD + S+K + +NW+ F + ++SA TI+VYIQD++G+ G GIPTI + ++I
Sbjct: 513 DQFDMTKKGVESRKWNLFNWYFFCMGFASLSALTIVVYIQDHMGWGWGLGIPTIAMFIAI 692
Query: 125 LVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEVSIEEYTSKGRYR 184
+ F+LG+ LY+ PSGSP R+ QV VAA RK +P D K L++ + + R
Sbjct: 693 IAFLLGSRLYKTLKPSGSPFLRLAQVIVAAFRKRNDALPNDPKLLYQNLELDSSISLEGR 872
Query: 185 INHSSSLRFLDKAAVKTGQTSP--------WMLCTVTQIEETKQMTKMLPVLITTCIPST 236
++H+ ++LDKAA+ T + + W L TV ++EE K + +MLP+ + + T
Sbjct: 873 LSHTDQYKWLDKAAIVTDEEAKNLNKLPNLWNLATVHRVEELKCLVRMLPIWASGILLIT 1052
Query: 237 IISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRVFVPVIRRYTKN 296
S + I Q T+DR + FEI PA + F + M+ V++Y+R+FVP IRR+TKN
Sbjct: 1053ASSSQHSFVIVQARTMDRHLSHTFEISPASMAIFSVLTMMTGVILYERLFVPFIRRFTKN 1232
Query: 297 PRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLL-GPLDTIPLTIFILVPQFA 355
P GIT LQR+GIG V+++I IV+ L+E KR VA + +LL P IP+++F LVPQ+
Sbjct: 1233PAGITCLQRMGIGFVINIIATIVSALLEIKRKKVASKYHLLDSPKAIIPISVFWLVPQYF 1412
Query: 356 LMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSIVADLTSKNGH 415
L G+A+ F+++ LEF YDQ+PESM+S T+ ++IG+F+ T L+++V T K
Sbjct: 1413LHGVAEVFMNVGHLEFLYDQSPESMRSSATALYCIAIAIGHFIGTLLVTLVHKYTGK--E 1586
Query: 416 KGWILN-NLNVSRIDYYYAFLALLSAVNFICFFVIAKFFVYNDDVTRTKMDLEMNPNS-- 472
+ W+ + NLN R++YYY + + +NFI + + A ++YN K E+N N+
Sbjct: 1587RNWLPDRNLNRGRLEYYYFLVCGIQVINFIYYVICA--WIYN-----YKPLEEINENNQG 1745
Query: 473 --SKHKAEISQVSYN 485
+ E+S VS N
Sbjct: 1746DLEQTNGELSLVSLN 1790
>AL387494 weakly similar to GP|17381202|gb putative peptide transporter
protein {Arabidopsis thaliana}, partial (25%)
Length = 495
Score = 260 bits (664), Expect = 9e-70
Identities = 124/164 (75%), Positives = 142/164 (85%)
Frame = +2
Query: 179 SKGRYRINHSSSLRFLDKAAVKTGQTSPWMLCTVTQIEETKQMTKMLPVLITTCIPSTII 238
S GR RI+ SSSL FLDKAA KTGQTSPWMLCTVTQ+EETKQMTKM+P+LITT IPST++
Sbjct: 2 SNGRNRIDRSSSLSFLDKAATKTGQTSPWMLCTVTQVEETKQMTKMIPILITTLIPSTLL 181
Query: 239 SQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRVFVPVIRRYTKNPR 298
Q+TTLF++QGTTLDR MG HF+IPPACL AF IFMLIS+V+YD FVP+IRRYTKNPR
Sbjct: 182 VQSTTLFVKQGTTLDRRMGPHFDIPPACLTAFTTIFMLISIVVYDLAFVPMIRRYTKNPR 361
Query: 299 GITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGPLDT 342
GIT+ QRLGIGLV+H+ +M+ ACL ERKRL VARENNL G DT
Sbjct: 362 GITLFQRLGIGLVLHIAIMVTACLAERKRLRVARENNLFGRHDT 493
>TC88300 similar to GP|9581817|emb|CAC00544.1 putative low-affinity nitrate
transporter {Nicotiana plumbaginifolia}, partial (56%)
Length = 1169
Score = 249 bits (636), Expect = 2e-66
Identities = 135/334 (40%), Positives = 206/334 (61%), Gaps = 11/334 (3%)
Frame = +1
Query: 135 RHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEVSI--EEYTSKGRYRINHSSSLR 192
R + +GSPLT++ V+VAA RK + +P DS L V +E K + + HS R
Sbjct: 40 RFKKLAGSPLTQIAVVYVAAWRKRNMELPYDSSLLFNVDDIEDEMLRKKKQVLPHSKQFR 219
Query: 193 FLDKAAVKTGQTS--------PWMLCTVTQIEETKQMTKMLPVLITTCIPSTIISQTTTL 244
FLDKAA+K +T W L T+T +EE K + +MLP+ TT + T+ +Q TT
Sbjct: 220 FLDKAAIKDPKTDGNEINVVRKWYLSTLTDVEEVKLVLRMLPIWATTIMFWTVYAQMTTF 399
Query: 245 FIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRVFVPVIRRYTKNPRGITMLQ 304
+ Q TTL+R +G F+IPPA L AF +L+++ IYDRV VP+ R+ KNP+G+T LQ
Sbjct: 400 SVSQATTLNRHIGKSFQIPPASLTAFFIGSILLTIPIYDRVIVPITRKIFKNPQGLTPLQ 579
Query: 305 RLGIGLVMHVIVMIVACLIERKRLSVARENNLL-GPLDTIPLTIFILVPQFALMGIADTF 363
R+G+GLV + M+ A L E KR+ +A +NL P IP+++F L+PQF +G + F
Sbjct: 580 RIGVGLVFSIFAMVAAALTELKRMRMAHLHNLTHNPNSEIPMSVFWLIPQFFFVGSGEAF 759
Query: 364 VDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSIVADLTSKNGHKGWILNNL 423
I +L+FF + P+ MK++ T +TLS+G F S+ L+++V +TS+ HK W+ +NL
Sbjct: 760 TYIGQLDFFLRECPKGMKTMSTGLFLSTLSLGFFFSSLLVTLVHKVTSQ--HKPWLADNL 933
Query: 424 NVSRIDYYYAFLALLSAVNFICFFVIAKFFVYND 457
N +++ +Y LALLS +N + + + A ++VY D
Sbjct: 934 NEAKLYNFYWLLALLSVLNLVIYLLCANWYVYKD 1035
>TC78128 weakly similar to GP|21928025|gb|AAM78041.1 At1g69870/T17F3_10
{Arabidopsis thaliana}, partial (77%)
Length = 2022
Score = 179 bits (453), Expect(2) = 3e-63
Identities = 100/287 (34%), Positives = 158/287 (54%), Gaps = 10/287 (3%)
Frame = +2
Query: 192 RFLDKAA-VKTGQTSP-------WMLCTVTQIEETKQMTKMLPVLITTCIPSTIISQTTT 243
R LDKAA V G +P W L ++ Q+EE K + + LP+ + T ++Q T
Sbjct: 1058 RVLDKAALVMEGDLNPDGTIVNQWNLVSIQQVEEVKCLARTLPIWAAGILGFTAMAQQGT 1237
Query: 244 LFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRVFVPVIRRYTKNPRGITML 303
+ Q +DR +G F+IP L I + + V YDR+ VP +R+ TKN GIT+L
Sbjct: 1238 FIVSQAMKMDRHLGPKFQIPAGSLGVISLIVIGLWVPFYDRICVPSLRKITKNEGGITLL 1417
Query: 304 QRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGPLDTIPLTIFILVPQFALMGIADTF 363
QR+GIG+V +I MIVA L+E+ R VA N P P+++ L PQ LMG + F
Sbjct: 1418 QRIGIGMVFSIISMIVAGLVEKVRRDVANSNPT--PQGIAPMSVMWLFPQLVLMGFCEAF 1591
Query: 364 VDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSIVADLTSKNGHKGWILNNL 423
I +EFF Q P+ M+S+ + + + ++ N++S+ L+ V +T + H W+ NN+
Sbjct: 1592 NIIGLIEFFNRQFPDHMRSIANALFSCSFALANYVSSILVITVHSVTKTHNHPDWLTNNI 1771
Query: 424 NVSRIDYYYAFLALLSAVNFICFFVIAKFFVYND--DVTRTKMDLEM 468
N R+DY+Y LA + +N + F +++ + Y D+ MD+E+
Sbjct: 1772 NEGRLDYFYYLLAGVGVLNLVYFLYVSQRYHYKGSVDIQEKPMDVEL 1912
Score = 81.6 bits (200), Expect(2) = 3e-63
Identities = 37/98 (37%), Positives = 60/98 (60%)
Frame = +3
Query: 62 MGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALGYGIPTILLV 121
+G DQFD K + SF+NW+ + + QT++VYIQD++ + G+ IPT+ ++
Sbjct: 663 LGVDQFDPTTEKGKKGINSFFNWYYTSFTVVLLFTQTVIVYIQDSISWKFGFAIPTLCML 842
Query: 122 VSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWK 159
SI++F +GT +Y H P GS + + QV VA+ +K K
Sbjct: 843 XSIILFFIGTKIYVHVKPEGSIFSSIAQVLVASFKKRK 956
>TC79608 similar to GP|20147231|gb|AAM10330.1 At1g68570/F24J5_7 {Arabidopsis
thaliana}, partial (59%)
Length = 1489
Score = 174 bits (441), Expect(2) = 1e-60
Identities = 100/265 (37%), Positives = 156/265 (58%), Gaps = 6/265 (2%)
Frame = +2
Query: 114 GIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEVS 173
GIPT+ +++SI+ F+ G PLYR+ P GSP TR+V+V VAA RK K+ +S L++
Sbjct: 5 GIPTLAMLISIIAFIGGYPLYRNLNPEGSPFTRLVKVGVAAFRKKKIPKVPNSTLLYQND 184
Query: 174 IEEYTSKGRYRINHSSSLRFLDKAAVKTGQTSP-----WMLCTVTQIEETKQMTKMLPVL 228
+ + ++ HS L+FLDKAA+ T + + W L TV ++EE K + +M P+
Sbjct: 185 ELDASITLGGKLLHSDQLKFLDKAAIVTEEDNTKTPDLWRLSTVHRVEELKSIIRMGPIW 364
Query: 229 ITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRVFVP 288
+ + T +Q T ++Q T++R + FEIP + F + ML + +YDRV +
Sbjct: 365 ASGILLITAYAQQGTFSLQQAKTMNRHLTKSFEIPAGSMSVFTILTMLFTTALYDRVLIR 544
Query: 289 VIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLL-GPLDTIPLTI 347
V RR+T RGIT L R+GIG V+ + VA +E KR VA E+ L+ + IP+++
Sbjct: 545 VARRFTGLDRGITFLHRMGIGFVISLFATFVAGFVEMKRKKVAMEHGLIEHSSEIIPISV 724
Query: 348 FILVPQFALMGIADTFVDIAKLEFF 372
F LVPQ++L G+A+ F+ I + F
Sbjct: 725 FWLVPQYSLHGMAEAFMSIGAFKSF 799
Score = 77.4 bits (189), Expect(2) = 1e-60
Identities = 35/87 (40%), Positives = 55/87 (62%), Gaps = 1/87 (1%)
Frame = +1
Query: 370 EFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSIVADLTSKNGHKGWIL-NNLNVSRI 428
EFFYDQAPESM S ++ T++S+GN++ST L+++V T W+ NNLN ++
Sbjct: 793 EFFYDQAPESMTSTAMAFFWTSISLGNYISTLLVTLVHKFTKGPNGTNWLPDNNLNKGKL 972
Query: 429 DYYYAFLALLSAVNFICFFVIAKFFVY 455
+Y+Y + LL +N I + + AK + Y
Sbjct: 973 EYFYWLITLLQFINLIYYLICAKMYTY 1053
>AL377677 weakly similar to GP|10716608|gb putative peptide transport protein
{Oryza sativa}, partial (18%)
Length = 447
Score = 218 bits (556), Expect = 3e-57
Identities = 106/149 (71%), Positives = 120/149 (80%)
Frame = +1
Query: 73 KERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALGYGIPTILLVVSILVFVLGTP 132
KER KLSF+NWW ILIG + + TILVYIQDNVG+ LGYG+PTI L VSI+ F +GTP
Sbjct: 1 KERFYKLSFFNWWFSSILIGMLFSSTILVYIQDNVGWGLGYGLPTIGLAVSIITFFVGTP 180
Query: 133 LYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEVSIEEYTSKGRYRINHSSSLR 192
YRHR PSGSP+TRM+QVFVAAMRKWK +VP D K LHE+SIEEY R +I H+S LR
Sbjct: 181 FYRHRFPSGSPITRMLQVFVAAMRKWKTHVPNDPKLLHELSIEEYACNARNKIEHTSFLR 360
Query: 193 FLDKAAVKTGQTSPWMLCTVTQIEETKQM 221
LDKAAV TGQTS WMLCTVTQ+EETKQM
Sbjct: 361 ILDKAAVTTGQTSSWMLCTVTQVEETKQM 447
>TC82302 similar to PIR|F84663|F84663 probable nitrate transporter
[imported] - Arabidopsis thaliana, partial (51%)
Length = 1133
Score = 213 bits (543), Expect = 1e-55
Identities = 120/335 (35%), Positives = 199/335 (58%), Gaps = 10/335 (2%)
Frame = +3
Query: 149 QVFVAAMRKWKLNVPIDSKELHEVSIEEYTSKGRYRINHSSSLRFLDKAAVKT------- 201
QV VA+++K K+ +P + L+E + E+ RI S RFL+KAA+
Sbjct: 9 QVIVASIKKRKMELPYNVGSLYEDTPEDS------RIEQSEQFRFLEKAAIVVEGDFDKD 170
Query: 202 ---GQTSPWMLCTVTQIEETKQMTKMLPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGA 258
+PW LC++T++EE K M ++LP+ TT I T +Q T + Q T++R +G
Sbjct: 171 LYGSGPNPWKLCSLTRVEEVKMMVRLLPIWATTIIFWTTYAQMITFSVEQAATMERNVG- 347
Query: 259 HFEIPPACLIAFVNIFMLISVVIYDRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMI 318
+F+IP L F + +L+++ + DR+ +P+ ++ P G + LQR IGL++ I M
Sbjct: 348 NFQIPAGSLTVFFVVAILLTLAVNDRIIMPLWKKLNGKP-GFSNLQRNAIGLLLSTIGMA 524
Query: 319 VACLIERKRLSVARENNLLGPLDTIPLTIFILVPQFALMGIADTFVDIAKLEFFYDQAPE 378
A LIE KRLSVA+ + G T+P+++F+LVPQF L+G + F+ +L+FF Q+P+
Sbjct: 525 AASLIEVKRLSVAK--GVKGNQTTLPISVFLLVPQFFLVGSGEAFIYTGQLDFFITQSPK 698
Query: 379 SMKSLGTSYATTTLSIGNFLSTFLLSIVADLTSKNGHKGWILNNLNVSRIDYYYAFLALL 438
MK++ T TTLS+G F+S+FL+S+V +T +GW+ +++N R+D +YA L +L
Sbjct: 699 GMKTMSTGLFLTTLSLGFFVSSFLVSVVKKVTGTRDGQGWLADHINKGRLDLFYALLTIL 878
Query: 439 SAVNFICFFVIAKFFVYNDDVTRTKMDLEMNPNSS 473
S +NF+ F V A ++ + K +++N +SS
Sbjct: 879 SFINFVAFLVCAFWY----KPKKPKPSMQINGSSS 971
>BE998753 similar to PIR|T45958|T45 oligopeptide transporter-like protein -
Arabidopsis thaliana, partial (44%)
Length = 809
Score = 210 bits (535), Expect = 9e-55
Identities = 109/262 (41%), Positives = 173/262 (65%), Gaps = 8/262 (3%)
Frame = +3
Query: 150 VFVAAMRKWKLNVPIDSKELHEVSIEEYTSKGRYRINHSSSLRFLDKAAVKTGQT----- 204
V VA++RK++++ P D L+E++ E KG +++H++ LRF DKAAV+ G++
Sbjct: 30 VIVASIRKYRVDAPTDKSLLYEIADTESAIKGSRKLDHTNELRFFDKAAVQ-GESDNLKE 206
Query: 205 --SPWMLCTVTQIEETKQMTKMLPVLITTCIPSTIISQTTTLFIRQGTTLDRGMG-AHFE 261
+PW LCTVTQ+EE K + ++LPV T I +T+ Q +TLF+ QG T++ +G + F+
Sbjct: 207 SINPWRLCTVTQVEELKSILRLLPVWATGIIFATVYGQMSTLFVLQGQTMNTHVGNSSFK 386
Query: 262 IPPACLIAFVNIFMLISVVIYDRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVAC 321
IPPA L F I ++ V +YDR+ VP+ R++T + G+T LQR+G+GL + + M+ A
Sbjct: 387 IPPASLSIFDTISVIFWVPVYDRIIVPIARKFTGHKNGLTQLQRMGVGLFISIFSMVAAA 566
Query: 322 LIERKRLSVARENNLLGPLDTIPLTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMK 381
+E RL R NN L+ IP+TIF VPQ+ L+G A+ F I +LEFFY+QAP++M+
Sbjct: 567 FLELVRLRTVRRNNYY-ELEEIPMTIFWQVPQYFLIGCAEVFTFIGQLEFFYEQAPDAMR 743
Query: 382 SLGTSYATTTLSIGNFLSTFLL 403
SL ++ + T++ G +LS+ L+
Sbjct: 744 SLCSALSLLTVAFGQYLSSLLV 809
>AW980440 similar to GP|17381202|gb putative peptide transporter protein
{Arabidopsis thaliana}, partial (23%)
Length = 688
Score = 201 bits (512), Expect = 4e-52
Identities = 95/140 (67%), Positives = 112/140 (79%)
Frame = +1
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G P STMGADQFD+F+ KE+S KLSF+NWW F ILIG + A T LVYIQDN+G+ LG
Sbjct: 127 GGTKPNISTMGADQFDDFDPKEKSDKLSFFNWWFFSILIGVLFATTFLVYIQDNIGWELG 306
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEV 172
YG+PTI L SILVF+LGTP YRH+LP GSP+TRM+QVFVAA+RKWK VP D KELHE+
Sbjct: 307 YGLPTIGLAFSILVFLLGTPYYRHKLPPGSPITRMLQVFVAAIRKWKARVPEDKKELHEL 486
Query: 173 SIEEYTSKGRYRINHSSSLR 192
S+EEYT GR RI+H+S R
Sbjct: 487 SMEEYTCNGRTRIDHTSFFR 546
>TC86049 weakly similar to GP|20466248|gb|AAM20441.1 putative transport
protein {Arabidopsis thaliana}, partial (43%)
Length = 1836
Score = 196 bits (497), Expect = 2e-50
Identities = 122/382 (31%), Positives = 201/382 (51%), Gaps = 14/382 (3%)
Frame = +2
Query: 81 FYNWWVFYILIGTISAQTILVYIQDNVGFALGYGIPTILLVVSILVFVLGTPLYRHRLPS 140
F++W+ + I I A T +VYIQD++G+ +G+G+P IL+++S ++F L +PLY
Sbjct: 110 FFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLISTVLFFLASPLYVKIKQK 289
Query: 141 GSPLTRMVQVFVAAMRKWKLNVPIDSKELHEVSIEEY-TSKGRYRINHSSSLRFLDKAAV 199
S T QV VAA + KL +P + S E Y K + + LRFL+KA V
Sbjct: 290 TSLFTGFAQVSVAAYKNRKLPLP------PKTSPEFYHQQKDSELVVPTDKLRFLNKACV 451
Query: 200 ----------KTGQTSPWMLCTVTQIEETKQMTKMLPVLITTCIPSTIISQTTTLFIRQG 249
+ W LCTV Q+EE K + K++P+ T S I + L Q
Sbjct: 452 IKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMSINIGGSFGLL--QA 625
Query: 250 TTLDRGM--GAHFEIPPACLIAFVNIFMLISVVIYDRVFVPVIRRYTKNPRGITMLQRLG 307
+LDR + ++FE+P + + +LI ++IYDRV +P+ + P I+ +R+G
Sbjct: 626 KSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMG 805
Query: 308 IGLVMHVIVMIVACLIER-KRLSVARENNLLGPLDTIPLTIFILVPQFALMGIADTFVDI 366
IGL + + +I A + E +R +E L + ++ L PQ L GIA+ F I
Sbjct: 806 IGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVI 985
Query: 367 AKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSIVADLTSKNGHKGWILNNLNVS 426
+ EF+Y + P+SM S+ S + + +GN +S+ +LSI+ T G++GW+ +N+N
Sbjct: 986 GQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKG 1165
Query: 427 RIDYYYAFLALLSAVNFICFFV 448
D YY + ++A+N + + V
Sbjct: 1166HFDKYYWVIVGINALNLLYYLV 1231
>TC79430 similar to PIR|F86358|F86358 Similar to peptide transporter
[imported] - Arabidopsis thaliana, partial (55%)
Length = 1303
Score = 195 bits (496), Expect = 3e-50
Identities = 100/280 (35%), Positives = 164/280 (57%), Gaps = 4/280 (1%)
Frame = +2
Query: 52 RGELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFAL 111
+G P GADQFD +ER + SF+NWW F G + +IL Y+QDNVG+ L
Sbjct: 479 QGGHKPCVQAFGADQFDINHPQERRSRSSFFNWWYFTFTAGLLVTVSILNYVQDNVGWVL 658
Query: 112 GYGIPTILLVVSILVFVLGTPLYRHRLPSGS---PLTRMVQVFVAAMRKWKLNVPIDSKE 168
G+GIP I +++++ +F LGT YR +P P +R+ +VF+ A+R ++
Sbjct: 659 GFGIPWIAMIIALSLFSLGTWTYRFSIPGNQQRGPFSRIGRVFITALRNFR--------- 811
Query: 169 LHEVSIEEYTSKGRYRINHSSSLRFLDKAAVKT-GQTSPWMLCTVTQIEETKQMTKMLPV 227
+ EE + S FL+KA + + G +C+V+++EE K + +++P+
Sbjct: 812 ----TTEEEEPRPSLLHQPSQQFSFLNKALIASDGSKENGKVCSVSEVEEAKAILRLVPI 979
Query: 228 LITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRVFV 287
T+ I + + SQ++T F +QG TLDR + F +PPA L +F+++ +++ + +YDR+ V
Sbjct: 980 WATSLIFAIVFSQSSTFFTKQGVTLDRKILPGFYVPPASLQSFISLSIVLFIPVYDRIIV 1159
Query: 288 PVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKR 327
P+ R +T P GITMLQR+G G++ VI M++A +E KR
Sbjct: 1160PIARTFTGKPSGITMLQRIGAGILFSVISMVIAAFVEMKR 1279
>TC84411 similar to GP|4102839|gb|AAD01600.1| LeOPT1 {Lycopersicon
esculentum}, partial (38%)
Length = 804
Score = 187 bits (476), Expect = 6e-48
Identities = 89/240 (37%), Positives = 157/240 (65%)
Frame = +1
Query: 214 QIEETKQMTKMLPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNI 273
Q+EE K + +M P+ T I S++ +Q +TLF+ QGT ++ +G+ F++ PA L F
Sbjct: 1 QVEELKILIRMFPIWATGIIFSSVYAQMSTLFVEQGTMMNTSIGS-FKLSPASLSTFEVA 177
Query: 274 FMLISVVIYDRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARE 333
+++ V +YD++ VP+++++T RGI++ QR+GIG + + M+ A +E KRL +ARE
Sbjct: 178 SVVMWVPVYDKILVPIVKKFTGKKRGISVFQRIGIGPFISGLCMLAAAAVEIKRLQLARE 357
Query: 334 NNLLGPLDTIPLTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLS 393
+L+ +PL++ +PQ+ ++G A+ F + +LEFFY+++P++M+++ + S
Sbjct: 358 LDLVDKPVGVPLSVLWQIPQYLILGAAEIFTFVGQLEFFYEESPDAMRTICGALPLLNFS 537
Query: 394 IGNFLSTFLLSIVADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFF 453
+GN+LS+F+L+IV T+K G GWI +NLN +DYY+ L+ LS +N + F V AK +
Sbjct: 538 LGNYLSSFILTIVTYFTTKGGRLGWIPDNLNKGHLDYYFLLLSGLSLLNMLVFIVAAKIY 717
>TC83116 similar to GP|11933407|dbj|BAB19758. putative nitrate transporter
NRT1-3 {Glycine max}, partial (50%)
Length = 1192
Score = 178 bits (452), Expect = 4e-45
Identities = 81/144 (56%), Positives = 111/144 (76%), Gaps = 2/144 (1%)
Frame = +3
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G P ST+GADQFD+F KE+S KLSF+NWW+F I GT+ A T+LVYIQDNVG+ LG
Sbjct: 531 GGTKPNISTIGADQFDDFHPKEKSHKLSFFNWWMFSIFFGTLFANTVLVYIQDNVGWTLG 710
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEV 172
Y +PT+ L +SI++F+ GTP YRH+LP+GS TRM +V VA++RKWK+ VP D+K+L+E+
Sbjct: 711 YALPTLGLAISIMIFLGGTPFYRHKLPAGSTFTRMARVIVASLRKWKVPVPDDTKKLYEL 890
Query: 173 SIEEYTSKG--RYRINHSSSLRFL 194
+EEY KG YRI+ + +LR++
Sbjct: 891 DMEEYAKKGSNTYRIDSTPTLRYI 962
>AW694862 similar to GP|10716600|gb putative peptide transport protein {Oryza
sativa}, partial (26%)
Length = 645
Score = 165 bits (418), Expect = 3e-41
Identities = 75/141 (53%), Positives = 106/141 (74%)
Frame = +1
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G P ST+GADQFD+F+ KE+S KLSF+NWW+ I+ G++ A T++VYIQDNVG+ LG
Sbjct: 142 GGTKPNISTIGADQFDDFDPKEKSLKLSFFNWWMSCIVFGSLFAFTVIVYIQDNVGWTLG 321
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEV 172
Y +PT+ L +SI+ F+ GTP YRH+L GSP M +V VAA+RK+ VP D KEL+E+
Sbjct: 322 YALPTLGLAISIITFLAGTPFYRHKLIKGSPFISMAKVIVAAIRKFDAVVPDDPKELYEL 501
Query: 173 SIEEYTSKGRYRINHSSSLRF 193
S+EEYT KG++RI+ + + ++
Sbjct: 502 SLEEYTKKGKFRIDLTQTFKY 564
>TC83960 weakly similar to GP|13605523|gb|AAK32755.1 AT3g53960/F5K20_260
{Arabidopsis thaliana}, partial (56%)
Length = 681
Score = 158 bits (400), Expect = 4e-39
Identities = 79/197 (40%), Positives = 121/197 (61%)
Frame = +3
Query: 261 EIPPACLIAFVNIFMLISVVIYDRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVA 320
++PPA + + M+ISV IYD++ VP++ + +N RGI +LQR+G G+ +I MIVA
Sbjct: 21 KLPPASIFTVAALGMIISVAIYDKILVPMLEKINQNERGINILQRIGFGMFFTIITMIVA 200
Query: 321 CLIERKRLSVARENNLLGPLDTIPLTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESM 380
LIE+KRL ++ PL+IF L PQF ++G D F + E+FYDQ P+SM
Sbjct: 201 ALIEKKRLEAVEKD---------PLSIFWLAPQFLIIGFGDGFTLVGLQEYFYDQVPDSM 353
Query: 381 KSLGTSYATTTLSIGNFLSTFLLSIVADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSA 440
+SLG ++ + + NF+S+ L++IV +T KNG K W +LN SR+D +Y LA ++
Sbjct: 354 RSLGIAFYLSVIGAANFISSVLITIVDHITKKNG-KSWFGKDLNSSRLDKFYWLLAAITT 530
Query: 441 VNFICFFVIAKFFVYND 457
VN F A+ + Y +
Sbjct: 531 VNLFLFVFFARRYSYKN 581
>TC84777 weakly similar to GP|20147231|gb|AAM10330.1 At1g68570/F24J5_7
{Arabidopsis thaliana}, partial (29%)
Length = 684
Score = 150 bits (380), Expect = 8e-37
Identities = 72/191 (37%), Positives = 119/191 (61%), Gaps = 1/191 (0%)
Frame = +3
Query: 207 WMLCTVTQIEETKQMTKMLPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPAC 266
W L TV ++EE K + ++LP+ + + + + I Q T+DR + F+I PA
Sbjct: 111 WKLATVHRVEELKSIIRILPISASGILLIAASAHLPSFVIEQARTMDRHLSHTFQISPAN 290
Query: 267 LIAFVNIFMLISVVIYDRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERK 326
+ F + M+ V++Y+RVF+P++R++T NP GIT +QR+GIG ++++I +++ +E K
Sbjct: 291 MSVFSVVTMMAGVILYERVFIPIVRKFTNNPVGITCIQRMGIGFIINIIATLISAPVEIK 470
Query: 327 RLSVARENNLL-GPLDTIPLTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGT 385
R VA + NLL P IP+++F LVPQ+ G+AD F+ + EF +DQAPESM S T
Sbjct: 471 RKEVAAKYNLLDDPKAIIPISVFWLVPQYCFHGLADVFMSVGLFEFLFDQAPESMXSTAT 650
Query: 386 SYATTTLSIGN 396
+ ++IG+
Sbjct: 651 AIYCIIIAIGS 683
>TC87895 similar to GP|20466248|gb|AAM20441.1 putative transport protein
{Arabidopsis thaliana}, partial (7%)
Length = 1265
Score = 142 bits (359), Expect = 2e-34
Identities = 83/278 (29%), Positives = 145/278 (51%), Gaps = 1/278 (0%)
Frame = +1
Query: 207 WMLCTVTQIEETKQMTKMLPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPAC 266
W LCTV Q+EE K + +++P+ T + S I + L Q +LDR + ++FE+P
Sbjct: 1 WRLCTVDQVEELKALVRVIPLWSTGIMMSLNIGGSFGLL--QAKSLDRHITSNFEVPAGS 174
Query: 267 LIAFVNIFMLISVVIYDRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERK 326
+ + I +V+YDRV +P+ + P I+ R+GIGL ++ + ++ A E
Sbjct: 175 FSVIMVGSIFIWIVLYDRVLIPLASKLRGKPVRISPKIRMGIGLFLNFLHLVTAAAFESI 354
Query: 327 RLSVARENNLLGPLD-TIPLTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGT 385
R A + L + ++ L PQ L GI++ F I + EF+Y + P +M S+
Sbjct: 355 RRKKAIQAGFLNDTHGVLKMSALWLAPQLCLGGISEAFNAIGQNEFYYKEFPRTMSSISA 534
Query: 386 SYATTTLSIGNFLSTFLLSIVADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFIC 445
S ++ GN +S+F+ + + ++TS+ G +GWI +N+N R D YY +A +SA+N +
Sbjct: 535 SLCGLGMAAGNLVSSFVFNTIENVTSRGGKEGWISDNINKGRYDKYYMVIAGVSALNLLY 714
Query: 446 FFVIAKFFVYNDDVTRTKMDLEMNPNSSKHKAEISQVS 483
F V + + Y V + + N + K E +V+
Sbjct: 715 FLVCS--WAYGPTVDQVSKVSDENDSKEKDSTEFKKVN 822
>TC79810 similar to GP|11994403|dbj|BAB02362. nitrate transporter
{Arabidopsis thaliana}, partial (60%)
Length = 1236
Score = 142 bits (357), Expect = 4e-34
Identities = 81/226 (35%), Positives = 130/226 (56%), Gaps = 1/226 (0%)
Frame = +3
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G + S G+DQFD + KE + F+N + F+I IG++ + +LVY+QDN+G G
Sbjct: 546 GGIKSNVSGFGSDQFDTNDPKEEKNMIFFFNRFYFFISIGSLFSVVVLVYVQDNIGRGWG 725
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEV 172
YGI ++V++ + + GTPLYR + P GSPLT + +V A +K L +P D L+
Sbjct: 726 YGISAGTMLVAVGILLCGTPLYRFKKPQGSPLTIIWRVLFLAWKKRTLPIPSDPTLLNGY 905
Query: 173 SIEEYTSKGRYRINHSSSLRFLDKAAVKTGQT-SPWMLCTVTQIEETKQMTKMLPVLITT 231
+ T ++R +++ LD+ K G +PW++ T+TQ+EE K + K+LP+ T
Sbjct: 906 LEAKVTYTDKFRSLDKAAI--LDETKSKDGNNENPWLVSTMTQVEEVKMVIKLLPIWSTC 1079
Query: 232 CIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLI 277
+ T+ SQ T I Q T ++R +G+ EIP L AF+ I +L+
Sbjct: 1080ILFWTVYSQMNTFTIEQATFMNRKVGS-LEIPAGSLSAFLFITILL 1214
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.140 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,686,228
Number of Sequences: 36976
Number of extensions: 189978
Number of successful extensions: 1666
Number of sequences better than 10.0: 84
Number of HSP's better than 10.0 without gapping: 1579
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1608
length of query: 485
length of database: 9,014,727
effective HSP length: 100
effective length of query: 385
effective length of database: 5,317,127
effective search space: 2047093895
effective search space used: 2047093895
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147517.7


