

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147517.11 + phase: 0
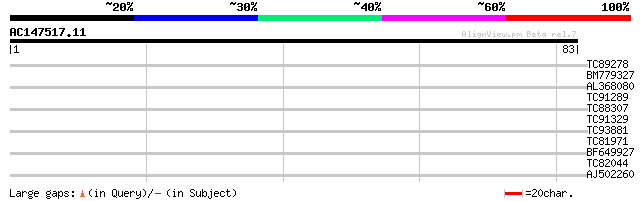
(83 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89278 similar to GP|9757719|dbj|BAB08244.1 CRN (crooked neck) ... 37 0.001
BM779327 weakly similar to GP|18700131|gb AT4g24270/T22A6_100 {A... 32 0.033
AL368080 similar to GP|20259567|gb unknown protein {Arabidopsis ... 32 0.044
TC91289 similar to GP|10177361|dbj|BAB10652. cell cycle control ... 31 0.097
TC88307 homologue to GP|6782273|emb|CAB70255.1 contains similari... 27 1.4
TC91329 similar to GP|7682783|gb|AAF67364.1| Hypothetical protei... 26 2.4
TC93881 similar to GP|9081779|dbj|BAA99518.1 hypothetical protei... 25 4.1
TC81971 similar to PIR|B96836|B96836 unknown protein T21F11.23 [... 25 4.1
BF649927 similar to GP|17065506|gb| Unknown protein {Arabidopsis... 25 5.3
TC82044 similar to PIR|B86285|B86285 hypothetical protein AAD396... 25 7.0
AJ502260 weakly similar to PIR|T51434|T514 hypothetical protein ... 24 9.1
>TC89278 similar to GP|9757719|dbj|BAB08244.1 CRN (crooked neck) protein
{Arabidopsis thaliana}, partial (64%)
Length = 1612
Score = 37.4 bits (85), Expect = 0.001
Identities = 22/81 (27%), Positives = 38/81 (46%), Gaps = 9/81 (11%)
Frame = +1
Query: 11 KMRRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYERAVQGVTYS---------V 61
K R Y+ + + PL Y W Y E +G+ ++ EVYERA+ V + +
Sbjct: 322 KRRFQYEDEVRKNPLNYDSWFDYIRLEESVGNKERTREVYERAIANVPPAEEKRYWQRYI 501
Query: 62 DMWLHYCIFAISTYGDPDTVR 82
+W++Y ++ GD + R
Sbjct: 502 YLWINYALYEELDAGDMERTR 564
Score = 32.3 bits (72), Expect = 0.033
Identities = 22/72 (30%), Positives = 34/72 (46%), Gaps = 3/72 (4%)
Frame = +1
Query: 2 IFLLKNNILKMRRVYDAFLAEFPL-CYGYWKKYADHEARLGSADKVVEVYERAVQGVTYS 60
I L NI + R++Y+ +L P CY W KYA+ E L ++ ++E A+
Sbjct: 742 IELQLGNIDRCRKLYEKYLEWSPENCYA-WSKYAELERSLAETERARAIFELAIAQPALD 918
Query: 61 VD--MWLHYCIF 70
+ MW Y F
Sbjct: 919 MPELMWKAYIDF 954
Score = 27.7 bits (60), Expect = 0.82
Identities = 18/55 (32%), Positives = 25/55 (44%)
Frame = +1
Query: 17 DAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLHYCIFA 71
D + L GY +YA E + G K VYERAV+ + + L + FA
Sbjct: 1 DLYCVILGLVLGY--RYAKFEMKNGEVPKARNVYERAVEKLADDEEAELLFVAFA 159
Score = 26.2 bits (56), Expect = 2.4
Identities = 11/38 (28%), Positives = 22/38 (56%)
Frame = +1
Query: 30 WKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLHY 67
+KKY + E +LG+ D+ ++YE+ ++ + W Y
Sbjct: 724 FKKYIEIELQLGNIDRCRKLYEKYLEWSPENCYAWSKY 837
>BM779327 weakly similar to GP|18700131|gb AT4g24270/T22A6_100 {Arabidopsis
thaliana}, partial (19%)
Length = 518
Score = 32.3 bits (72), Expect = 0.033
Identities = 14/60 (23%), Positives = 31/60 (51%), Gaps = 1/60 (1%)
Frame = +3
Query: 11 KMRRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVV-EVYERAVQGVTYSVDMWLHYCI 69
+++ +Y+ + +FPL W Y + + K+V +VY RA + + ++W+ Y +
Sbjct: 231 RIQVLYERAITDFPLSPDLWLDYTHYLDKTLKVGKIVSDVYSRATKNCPWVGELWVRYML 410
Score = 28.5 bits (62), Expect = 0.48
Identities = 12/38 (31%), Positives = 19/38 (49%)
Frame = +3
Query: 30 WKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLHY 67
+ Y E G+ ++ +YERA+ S D+WL Y
Sbjct: 186 YMNYLKFEQSSGTPARIQVLYERAITDFPLSPDLWLDY 299
>AL368080 similar to GP|20259567|gb unknown protein {Arabidopsis thaliana},
partial (15%)
Length = 389
Score = 32.0 bits (71), Expect = 0.044
Identities = 13/40 (32%), Positives = 19/40 (47%)
Frame = +3
Query: 30 WKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLHYCI 69
W Y D R G K+V++YER V + W+ Y +
Sbjct: 204 WHNYLDFIEREGDLSKIVKLYERCVIACANYPEYWIRYVL 323
>TC91289 similar to GP|10177361|dbj|BAB10652. cell cycle control crn
(crooked neck) protein-like {Arabidopsis thaliana},
partial (20%)
Length = 698
Score = 30.8 bits (68), Expect = 0.097
Identities = 14/48 (29%), Positives = 24/48 (49%), Gaps = 2/48 (4%)
Frame = +1
Query: 30 WKKYADHEARLGSADKVVEVYERAVQGVTYSVD--MWLHYCIFAISTY 75
W Y + +G+ ++ +EVYERA+ V + + W HY + Y
Sbjct: 331 WFDYIRWKESVGNKERTMEVYERAIANVPLADENRYWQHYIYLCYALY 474
>TC88307 homologue to GP|6782273|emb|CAB70255.1 contains similarity to Pfam
domain: PF01748 (Domain of unknown function) Score=122.0
E-value=3., partial (4%)
Length = 1681
Score = 26.9 bits (58), Expect = 1.4
Identities = 8/27 (29%), Positives = 18/27 (66%)
Frame = +2
Query: 7 NNILKMRRVYDAFLAEFPLCYGYWKKY 33
+N+L ++ + + E+PL +GY+ +Y
Sbjct: 1484 SNLLSLKMISNEISTEYPLYFGYYPRY 1564
>TC91329 similar to GP|7682783|gb|AAF67364.1| Hypothetical protein T32B20.g
{Arabidopsis thaliana}, partial (38%)
Length = 1068
Score = 26.2 bits (56), Expect = 2.4
Identities = 13/41 (31%), Positives = 20/41 (48%)
Frame = +3
Query: 30 WKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLHYCIF 70
W + D E LGS + EVYER + + + ++Y F
Sbjct: 309 WTFFVDLEESLGSLESTREVYERILDLRIATPQIIINYAYF 431
>TC93881 similar to GP|9081779|dbj|BAA99518.1 hypothetical protein {Oryza
sativa (japonica cultivar-group)}, partial (27%)
Length = 826
Score = 25.4 bits (54), Expect = 4.1
Identities = 9/31 (29%), Positives = 18/31 (58%)
Frame = -2
Query: 25 LCYGYWKKYADHEARLGSADKVVEVYERAVQ 55
LC G +A HE++LGS + + ++ ++
Sbjct: 294 LCLGVSNGFATHESKLGSPAPGIRILDKPIE 202
>TC81971 similar to PIR|B96836|B96836 unknown protein T21F11.23 [imported] -
Arabidopsis thaliana, partial (13%)
Length = 721
Score = 25.4 bits (54), Expect = 4.1
Identities = 17/57 (29%), Positives = 27/57 (46%), Gaps = 2/57 (3%)
Frame = +1
Query: 28 GYWKKYADHEARLGSADK--VVEVYERAVQGVTYSVDMWLHYCIFAISTYGDPDTVR 82
G W K + E G ++E+ E + YSV+M H+C +S + P T+R
Sbjct: 517 GVWSKLENPENFTGHVQSGCLLEI*EFCFVLIYYSVNMKQHFCFLLLSYF--P*TLR 681
>BF649927 similar to GP|17065506|gb| Unknown protein {Arabidopsis thaliana},
partial (8%)
Length = 633
Score = 25.0 bits (53), Expect = 5.3
Identities = 11/27 (40%), Positives = 16/27 (58%)
Frame = -1
Query: 4 LLKNNILKMRRVYDAFLAEFPLCYGYW 30
+LKN + R + F+ EFP YG+W
Sbjct: 507 VLKNFVNNNPR-FSLFIGEFPSLYGWW 430
>TC82044 similar to PIR|B86285|B86285 hypothetical protein AAD39642.1
[imported] - Arabidopsis thaliana, partial (16%)
Length = 950
Score = 24.6 bits (52), Expect = 7.0
Identities = 10/25 (40%), Positives = 14/25 (56%)
Frame = -2
Query: 44 DKVVEVYERAVQGVTYSVDMWLHYC 68
D +E Y+ A V V +W+HYC
Sbjct: 796 DMEMEWYKEA*DQVVAVVGVWVHYC 722
>AJ502260 weakly similar to PIR|T51434|T514 hypothetical protein F2G14_10 -
Arabidopsis thaliana, partial (3%)
Length = 667
Score = 24.3 bits (51), Expect = 9.1
Identities = 8/20 (40%), Positives = 14/20 (70%)
Frame = -1
Query: 61 VDMWLHYCIFAISTYGDPDT 80
+D++LH+C+ IST+ T
Sbjct: 415 IDLYLHHCLAPISTHKHEST 356
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.328 0.142 0.456
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,725,168
Number of Sequences: 36976
Number of extensions: 27602
Number of successful extensions: 278
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 269
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 278
length of query: 83
length of database: 9,014,727
effective HSP length: 59
effective length of query: 24
effective length of database: 6,833,143
effective search space: 163995432
effective search space used: 163995432
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 51 (24.3 bits)
Medicago: description of AC147517.11


