

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
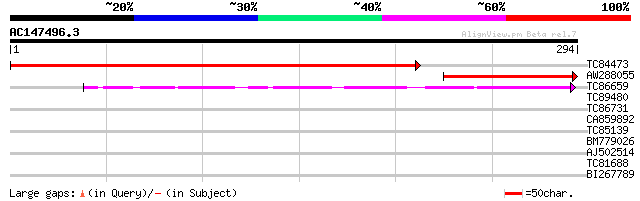
Query= AC147496.3 - phase: 0
(294 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC84473 similar to GP|23095415|emb|CAD46621. unknown {Streptococ... 425 e-120
AW288055 155 1e-38
TC86659 similar to GP|5360263|dbj|BAA81904.1 NtPRp27 {Nicotiana ... 66 1e-11
TC89480 similar to GP|20260684|gb|AAM13240.1 unknown protein {Ar... 28 3.2
TC86731 similar to GP|5295968|dbj|BAA81869.1 ESTs C19441(E10423)... 28 3.2
CA859892 similar to GP|23510639|em hypothetical protein {Plasmod... 28 4.2
TC85139 homologue to GP|13540318|gb|AAK29410.1 S-adenosyl-L-meth... 27 9.4
BM779026 homologue to GP|13540318|gb S-adenosyl-L-methionine syn... 27 9.4
AJ502514 similar to GP|23496962|gb| hypothetical protein {Plasmo... 27 9.4
TC81688 weakly similar to PIR|T51471|T51471 farnesylated protein... 27 9.4
BI267789 27 9.4
>TC84473 similar to GP|23095415|emb|CAD46621. unknown {Streptococcus
agalactiae}, partial (2%)
Length = 756
Score = 425 bits (1093), Expect = e-120
Identities = 212/213 (99%), Positives = 212/213 (99%)
Frame = -1
Query: 1 MENEDKHFLFQPLLPHFNPSSYEINQQNDYNNSKFFIPNNNNIIHRILLILFVAIISIWA 60
MENEDKHFLFQPLLPHFNPSSYEINQQNDYNNSKFFIPNNNNIIHRILLILFVAIISIWA
Sbjct: 639 MENEDKHFLFQPLLPHFNPSSYEINQQNDYNNSKFFIPNNNNIIHRILLILFVAIISIWA 460
Query: 61 NYEASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRILLNTSSFVEQILYPNGNNDNID 120
NYEASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRILLNTSSFVEQILYPNGNNDNID
Sbjct: 459 NYEASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRILLNTSSFVEQILYPNGNNDNID 280
Query: 121 IKNKKIIKSVTLRLARQNLDTITTITADEKNINSYVIEISPMLLEDENFNNMAIVGAIQR 180
IKNKKIIKSVTLRLARQNLDTITTITADEKNINSYVIEISPMLLEDENFNNMAIVGAIQR
Sbjct: 279 IKNKKIIKSVTLRLARQNLDTITTITADEKNINSYVIEISPMLLEDENFNNMAIVGAIQR 100
Query: 181 AMARVWLWDGRSKASPRLLDGMVEYIAELAGFH 213
AMARVWLWDGRSKASP LLDGMVEYIAELAGFH
Sbjct: 99 AMARVWLWDGRSKASPXLLDGMVEYIAELAGFH 1
>AW288055
Length = 375
Score = 155 bits (393), Expect = 1e-38
Identities = 69/69 (100%), Positives = 69/69 (100%)
Frame = +3
Query: 226 ECEDGRELWWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVGDVLGLKATKL 285
ECEDGRELWWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVGDVLGLKATKL
Sbjct: 3 ECEDGRELWWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVGDVLGLKATKL 182
Query: 286 CGLYNATYL 294
CGLYNATYL
Sbjct: 183 CGLYNATYL 209
>TC86659 similar to GP|5360263|dbj|BAA81904.1 NtPRp27 {Nicotiana tabacum},
partial (83%)
Length = 875
Score = 66.2 bits (160), Expect = 1e-11
Identities = 59/255 (23%), Positives = 111/255 (43%)
Frame = +1
Query: 39 NNNNIIHRILLILFVAIISIWANYEASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRI 98
N N I+H L F+ +++I +A D + N+ + G RF + A++
Sbjct: 43 NTNTIMHT--LYFFLVLLAIIHGIQA---VDYSVTNNALSTPGGVRFR-DQLGAQYATQT 204
Query: 99 LLNTSSFVEQILYPNGNNDNIDIKNKKIIKSVTLRLARQNLDTITTITADEKNINSYVIE 158
L + + F+ ++ N D +K ++ V+L + ++D + + +E ++++ +
Sbjct: 205 LDSATQFIWRVFQQNNPAD------RKNVQKVSLFV--DDMDGVAYTSNNEIHLSARYVN 360
Query: 159 ISPMLLEDENFNNMAIVGAIQRAMARVWLWDGRSKASPRLLDGMVEYIAELAGFHRERLS 218
L E I G + M VW W+G +A+ L++G+ +Y+ RL
Sbjct: 361 SYGGDLRKE------ITGVLYHEMTHVWQWNGNGQANGGLIEGIADYV---------RLK 495
Query: 219 GGVGESPECEDGRELWWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVGDVL 278
S + G+ WD AR L YC+ GF+ LN+ M++ + D+ +L
Sbjct: 496 ANYAPSHWVKPGQGNKWDQGYDV-TARFLDYCDTLRSGFVAELNKLMRNGYSDQFFVQLL 672
Query: 279 GLKATKLCGLYNATY 293
G +L Y A Y
Sbjct: 673 GKTVNQLWTEYKAKY 717
>TC89480 similar to GP|20260684|gb|AAM13240.1 unknown protein {Arabidopsis
thaliana}, partial (43%)
Length = 1068
Score = 28.5 bits (62), Expect = 3.2
Identities = 17/45 (37%), Positives = 24/45 (52%), Gaps = 8/45 (17%)
Frame = +2
Query: 11 QPLLP--------HFNPSSYEINQQNDYNNSKFFIPNNNNIIHRI 47
QPL+P + NP +I N NN+ FFI NNNN+ + +
Sbjct: 92 QPLIPIPKFNLNLNLNPIKTQIFIFN--NNNNFFINNNNNLFNNL 220
>TC86731 similar to GP|5295968|dbj|BAA81869.1 ESTs C19441(E10423)
C99201(E10423) correspond to a region of the predicted
gene.~hypothetical, partial (52%)
Length = 1560
Score = 28.5 bits (62), Expect = 3.2
Identities = 14/39 (35%), Positives = 21/39 (52%)
Frame = -3
Query: 39 NNNNIIHRILLILFVAIISIWANYEASKTFDIHIVNDTK 77
++N HR +++ F+AII+ Y K H NDTK
Sbjct: 1474 DHNLDTHRTMVLHFIAIITWTRGYIKQKIIKRHTYNDTK 1358
>CA859892 similar to GP|23510639|em hypothetical protein {Plasmodium
falciparum 3D7}, partial (1%)
Length = 459
Score = 28.1 bits (61), Expect = 4.2
Identities = 34/119 (28%), Positives = 48/119 (39%), Gaps = 5/119 (4%)
Frame = +1
Query: 5 DKHFLFQP-LLPHFNPSSYEIN----QQNDYNNSKFFIPNNNNIIHRILLILFVAIISIW 59
+KH L+ L +PSSY+ N N+ NN K NNNN + L
Sbjct: 166 EKHNLYNNNTLLSSSPSSYKANLLLFSNNNDNNDK----NNNNDDYGATPPL-------- 309
Query: 60 ANYEASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRILLNTSSFVEQILYPNGNNDN 118
+S TF IH ++ K S +R FY ++D ++ Y N NDN
Sbjct: 310 --SPSSPTFPIHHHHNNKVSYGRKRLLDFYDNDD------------IDDDYYNNNINDN 444
>TC85139 homologue to GP|13540318|gb|AAK29410.1 S-adenosyl-L-methionine
synthetase {Elaeagnus umbellata}, complete
Length = 1542
Score = 26.9 bits (58), Expect = 9.4
Identities = 16/55 (29%), Positives = 31/55 (56%), Gaps = 1/55 (1%)
Frame = -3
Query: 91 SNDKASRILLNTSSFVEQILYPNGNNDNIDIK-NKKIIKSVTLRLARQNLDTITT 144
+NDKASR + + Q+L N DN+ +K + ++ + +L + +N D++ T
Sbjct: 835 TNDKASRRVKVEDCLLIQVLLGNNRLDNMFLKISSNLVVTDSLVVLSRNEDSVDT 671
>BM779026 homologue to GP|13540318|gb S-adenosyl-L-methionine synthetase
{Elaeagnus umbellata}, partial (69%)
Length = 800
Score = 26.9 bits (58), Expect = 9.4
Identities = 16/55 (29%), Positives = 31/55 (56%), Gaps = 1/55 (1%)
Frame = -1
Query: 91 SNDKASRILLNTSSFVEQILYPNGNNDNIDIK-NKKIIKSVTLRLARQNLDTITT 144
+NDKASR + + Q+L N DN+ +K + ++ + +L + +N D++ T
Sbjct: 494 TNDKASRRVKVEDCLLIQVLLGNNRLDNMFLKISSNLVVTDSLVVLSRNEDSVDT 330
>AJ502514 similar to GP|23496962|gb| hypothetical protein {Plasmodium
falciparum 3D7}, partial (9%)
Length = 417
Score = 26.9 bits (58), Expect = 9.4
Identities = 14/60 (23%), Positives = 25/60 (41%)
Frame = +3
Query: 91 SNDKASRILLNTSSFVEQILYPNGNNDNIDIKNKKIIKSVTLRLARQNLDTITTITADEK 150
+N+K+S T++ Q YPN ++ K+ K K N+D + +K
Sbjct: 72 ANNKSSSTTTATTTVTTQAAYPNSQQSHLSSKDNKQAKEKEKEKETTNVDDDQAVEEPQK 251
>TC81688 weakly similar to PIR|T51471|T51471 farnesylated protein ATFP6-like
protein - Arabidopsis thaliana, partial (44%)
Length = 697
Score = 26.9 bits (58), Expect = 9.4
Identities = 16/45 (35%), Positives = 21/45 (46%)
Frame = -3
Query: 24 INQQNDYNNSKFFIPNNNNIIHRILLILFVAIISIWANYEASKTF 68
IN N +N FF+ +NN I H L L + + Y KTF
Sbjct: 665 INFTNQFNTF-FFLSSNNYINHNTLFFLSFFLFILRCIYTLHKTF 534
>BI267789
Length = 661
Score = 26.9 bits (58), Expect = 9.4
Identities = 19/65 (29%), Positives = 30/65 (45%), Gaps = 2/65 (3%)
Frame = +3
Query: 103 SSFVEQILYPNGNNDNIDIKNKKIIK--SVTLRLARQNLDTITTITADEKNINSYVIEIS 160
SSF + L + D D+ NKK+I +TL Q LD T +N+ +++ +
Sbjct: 225 SSFFQVTLDRIASRDFKDLFNKKLINKLEITLNSINQLLDDAETKKYQNQNVKNWLDRLK 404
Query: 161 PMLLE 165
L E
Sbjct: 405 HKLYE 419
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.137 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,359,978
Number of Sequences: 36976
Number of extensions: 135162
Number of successful extensions: 811
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 800
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 809
length of query: 294
length of database: 9,014,727
effective HSP length: 95
effective length of query: 199
effective length of database: 5,502,007
effective search space: 1094899393
effective search space used: 1094899393
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC147496.3


