

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147483.4 + phase: 0
(310 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
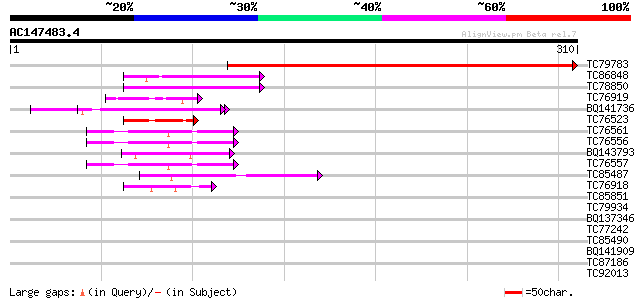
Score E
Sequences producing significant alignments: (bits) Value
TC79783 weakly similar to PIR|E96576|E96576 unknown protein 435... 387 e-108
TC86848 similar to GP|10177413|dbj|BAB10544. fibrillarin-like {A... 51 5e-07
TC78850 homologue to GP|10177413|dbj|BAB10544. fibrillarin-like ... 48 5e-06
TC76919 similar to GP|15724324|gb|AAL06555.1 At1g76010/T4O12_22 ... 44 1e-04
BQ141736 similar to PIR|T06291|T062 extensin homolog T9E8.80 - A... 44 1e-04
TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 42 3e-04
TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 41 5e-04
TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 41 5e-04
BQ143793 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 41 5e-04
TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 41 5e-04
TC85487 homologue to SP|P49688|RS2_ARATH 40S ribosomal protein S... 40 9e-04
TC76918 similar to GP|2231329|gb|AAB62000.1| bactinecin 11 {Ovis... 40 9e-04
TC85851 similar to PIR|T12180|T12180 probable transcription fact... 39 0.002
TC79934 similar to GP|20259633|gb|AAM14173.1 putative GAR1 prote... 39 0.002
BQ137346 homologue to GP|15027095|em hypothetical protein LT.09 ... 39 0.003
TC77242 similar to GP|22655123|gb|AAM98152.1 putative protein {A... 39 0.003
TC85490 similar to GP|18146720|dbj|BAB82426. ribosomal protein S... 39 0.003
BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox ... 38 0.004
TC87186 similar to PIR|F96687|F96687 hypothetical protein T6J19.... 37 0.007
TC92013 homologue to GP|11595557|emb|CAC18142. related to c-modu... 37 0.007
>TC79783 weakly similar to PIR|E96576|E96576 unknown protein 43598-45751
[imported] - Arabidopsis thaliana, partial (29%)
Length = 1050
Score = 387 bits (993), Expect = e-108
Identities = 191/191 (100%), Positives = 191/191 (100%)
Frame = +2
Query: 120 DGLYVGDNADGEKLAKKLGPEIMDQITEAYEEIIERVLPSPLQDEYVEAMDINCAIEFEP 179
DGLYVGDNADGEKLAKKLGPEIMDQITEAYEEIIERVLPSPLQDEYVEAMDINCAIEFEP
Sbjct: 53 DGLYVGDNADGEKLAKKLGPEIMDQITEAYEEIIERVLPSPLQDEYVEAMDINCAIEFEP 232
Query: 180 EYAVEFDNPDIDEKEPIALRDALEKMKPFLMTYEGIRSQEEWEEVIEELMQRVPLLKKIV 239
EYAVEFDNPDIDEKEPIALRDALEKMKPFLMTYEGIRSQEEWEEVIEELMQRVPLLKKIV
Sbjct: 233 EYAVEFDNPDIDEKEPIALRDALEKMKPFLMTYEGIRSQEEWEEVIEELMQRVPLLKKIV 412
Query: 240 DHYSGPDRVTAKKQQEELERVAKTLPTSAPSSVKEFTNRAVVSLQSNPGWGFDKKCQFMD 299
DHYSGPDRVTAKKQQEELERVAKTLPTSAPSSVKEFTNRAVVSLQSNPGWGFDKKCQFMD
Sbjct: 413 DHYSGPDRVTAKKQQEELERVAKTLPTSAPSSVKEFTNRAVVSLQSNPGWGFDKKCQFMD 592
Query: 300 KLVFEVSQHHK 310
KLVFEVSQHHK
Sbjct: 593 KLVFEVSQHHK 625
>TC86848 similar to GP|10177413|dbj|BAB10544. fibrillarin-like {Arabidopsis
thaliana}, partial (92%)
Length = 1250
Score = 51.2 bits (121), Expect = 5e-07
Identities = 33/85 (38%), Positives = 41/85 (47%), Gaps = 8/85 (9%)
Frame = +3
Query: 63 GDGSGRGRGRG--------RGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDI 114
GD GRG GRG RG ARG G RG GGRGGRG GRGG + G S +
Sbjct: 111 GDRGGRGGGRGGFGGRGGDRGTPFKARG-GGRGGGGRGGRGGGRGGGRGGGMKGGSKVIV 287
Query: 115 ARSNADGLYVGDNADGEKLAKKLGP 139
+G+++ + + K L P
Sbjct: 288 EPHRHEGIFIAKGKEDALVTKNLVP 362
Score = 34.3 bits (77), Expect = 0.063
Identities = 26/51 (50%), Positives = 26/51 (50%), Gaps = 1/51 (1%)
Frame = +3
Query: 59 VRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRG-GFKRYGDDR 108
VR G G RG GRG D RG GRGG GGRG RG FK G R
Sbjct: 69 VRGRGGGGFRG---GRGGD---RGGRGGGRGGFGGRGGDRGTPFKARGGGR 203
>TC78850 homologue to GP|10177413|dbj|BAB10544. fibrillarin-like
{Arabidopsis thaliana}, partial (87%)
Length = 1177
Score = 47.8 bits (112), Expect = 5e-06
Identities = 30/78 (38%), Positives = 37/78 (46%), Gaps = 1/78 (1%)
Frame = +3
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRG-GFKRYGDDRTSHQDIARSNADG 121
G G GRG G GRG D RG G GGRG RG GRG G R G S + +G
Sbjct: 108 GGGGGRGFGGGRGGDFKPRGGGRGFGGGRGARGGGRGRGGGRGGMKGGSKVVVEPHRHEG 287
Query: 122 LYVGDNADGEKLAKKLGP 139
+++ + + K L P
Sbjct: 288 IFIAKGKEDALVTKNLVP 341
Score = 32.0 bits (71), Expect = 0.31
Identities = 22/45 (48%), Positives = 24/45 (52%), Gaps = 3/45 (6%)
Frame = +3
Query: 67 GRGRGRGRGRDVYARGRGDRGRG---GRGGRGDGRGGFKRYGDDR 108
GRG GRG D RG G GRG GRGG RGG + +G R
Sbjct: 63 GRGGFGGRGGDRGGRGGGG-GRGFGGGRGGDFKPRGGGRGFGGGR 194
Score = 28.9 bits (63), Expect = 2.6
Identities = 12/15 (80%), Positives = 12/15 (80%)
Frame = +3
Query: 86 RGRGGRGGRGDGRGG 100
RGRGG GGRG RGG
Sbjct: 60 RGRGGFGGRGGDRGG 104
Score = 27.3 bits (59), Expect = 7.7
Identities = 14/20 (70%), Positives = 14/20 (70%)
Frame = +3
Query: 81 RGRGDRGRGGRGGRGDGRGG 100
RGRG G GGRGG GRGG
Sbjct: 60 RGRG--GFGGRGGDRGGRGG 113
>TC76919 similar to GP|15724324|gb|AAL06555.1 At1g76010/T4O12_22
{Arabidopsis thaliana}, partial (42%)
Length = 1207
Score = 43.5 bits (101), Expect = 1e-04
Identities = 29/56 (51%), Positives = 34/56 (59%), Gaps = 3/56 (5%)
Frame = +3
Query: 53 QPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGG---RGDGRGGFKRYG 105
+P+N + D+G+GS R RGRGRGR RGRG RGRG G GDG G YG
Sbjct: 522 KPLNEY-EDEGEGSPRMRGRGRGR---GRGRG-RGRGMYNGGMEYGDGWDGGPGYG 674
Score = 37.4 bits (85), Expect = 0.007
Identities = 21/41 (51%), Positives = 22/41 (53%)
Frame = +3
Query: 59 VRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRG 99
+R G G GRGRGRGRG GD GG G G GRG
Sbjct: 567 MRGRGRGRGRGRGRGRGMYNGGMEYGDGWDGGPGYGGRGRG 689
Score = 35.8 bits (81), Expect = 0.022
Identities = 23/54 (42%), Positives = 25/54 (45%)
Frame = +3
Query: 45 ESDNVPRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGR 98
E + PR + R G G GRGRGRG G G G G GGRG GR
Sbjct: 546 EGEGSPRMRGRGR-----GRGRGRGRGRGMYNGGMEYGDGWDGGPGYGGRGRGR 692
Score = 31.6 bits (70), Expect = 0.41
Identities = 23/59 (38%), Positives = 23/59 (38%), Gaps = 20/59 (33%)
Frame = +3
Query: 61 DDGDG-SGRGRGRGRGRDVYARGRG-------------------DRGRGGRGGRGDGRG 99
D G G GRGRGR G RGRG RGRG GR GRG
Sbjct: 654 DGGPGYGGRGRGRAWGHAFRGRGRGYGAQPVGQYDYGEYDAPPAPRGRGRERGRARGRG 830
Score = 28.1 bits (61), Expect = 4.5
Identities = 18/38 (47%), Positives = 22/38 (57%), Gaps = 1/38 (2%)
Frame = +3
Query: 53 QPMNRFVRDDGDGSGRGRGRGRGRDVYARGRG-DRGRG 89
QP+ ++ + D RGRGR R ARGRG D GRG
Sbjct: 738 QPVGQYDYGEYDAPPAPRGRGRERG-RARGRGRDAGRG 848
>BQ141736 similar to PIR|T06291|T062 extensin homolog T9E8.80 - Arabidopsis
thaliana, partial (4%)
Length = 1099
Score = 43.5 bits (101), Expect = 1e-04
Identities = 35/115 (30%), Positives = 51/115 (43%), Gaps = 6/115 (5%)
Frame = +1
Query: 12 RGRGKPLEEAAQEAPQAPVVNRHIRVR-----QTPADAESDNVPRRQPMNRFVRDDGDGS 66
R R +P Q P ++H + + +T + P+ +P R R+ G
Sbjct: 148 RTRPRPTPPRPTADLQQPQADKHTKPKHASKGETKGKEKKFTAPQTRPA-RPAREGGPRR 324
Query: 67 GRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDR-TSHQDIARSNAD 120
G GRGRGRG D + + GGRGG G +GG GD+ + +D R AD
Sbjct: 325 G-GRGRGRGPDDPQERQTETENGGRGGNGGRKGGSDGGGDENGGAEKDNRRRPAD 486
Score = 40.4 bits (93), Expect = 9e-04
Identities = 27/82 (32%), Positives = 40/82 (47%), Gaps = 1/82 (1%)
Frame = +1
Query: 38 RQTPADAESDNVPRRQ-PMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGD 96
R+ PAD PR++ P N+ R G + GR + RGRGD R RG RG
Sbjct: 469 RRRPADPR----PRKEKPTNKRKRGTGTKKPKREGRRGEKKGRERGRGDGNREKRGERGG 636
Query: 97 GRGGFKRYGDDRTSHQDIARSN 118
G+G K+ + R + ++ R +
Sbjct: 637 GKGRTKKEKEKRPTKEEERREH 702
Score = 29.3 bits (64), Expect = 2.0
Identities = 26/110 (23%), Positives = 38/110 (33%), Gaps = 5/110 (4%)
Frame = +3
Query: 4 PKVLPGGGRGRGKPLEEAAQEAPQAPVVNRHIRVRQTPADAESDNVPRRQPM-----NRF 58
P PG G G G+P + + + P R RQ RQP
Sbjct: 318 PAGRPGEGEGTGRPARKTDGDRERRPGGERGPERRQRRGRGRKRGGGERQPAATGRPQAE 497
Query: 59 VRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDR 108
R + G G + + +G +R GGR G G +R G+ +
Sbjct: 498 KRKTNEQKKAGHGHQKTKKRGPQGGEEREGKRPGGRKQGEAGRERGGEGK 647
>TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (69%)
Length = 1615
Score = 42.0 bits (97), Expect = 3e-04
Identities = 24/41 (58%), Positives = 25/41 (60%)
Frame = +3
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKR 103
G GR GRG GR GR GRGGRGGRG GRGGF +
Sbjct: 1173 GGFGGRSGGRGGGR---FGGRDSGGRGGRGGRG-GRGGFNK 1283
Score = 33.5 bits (75), Expect = 0.11
Identities = 23/46 (50%), Positives = 24/46 (52%)
Frame = +3
Query: 62 DGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDD 107
D GSG GRG G GR R G GG GGR GRGG + G D
Sbjct: 1098 DSQGSG-GRGGG-GRSGGGRFGGGGRSGGFGGRSGGRGGGRFGGRD 1229
Score = 29.3 bits (64), Expect = 2.0
Identities = 19/47 (40%), Positives = 21/47 (44%), Gaps = 1/47 (2%)
Frame = +3
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRG-GRGDGRGGFKRYG 105
RD GRG G G + G G GGR GRG GR G + G
Sbjct: 1095 RDSQGSGGRGGGGRSGGGRFGGGGRSGGFGGRSGGRGGGRFGGRDSG 1235
>TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 1054
Score = 41.2 bits (95), Expect = 5e-04
Identities = 32/88 (36%), Positives = 37/88 (41%), Gaps = 5/88 (5%)
Frame = +1
Query: 43 DAESDNVPRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGD-----RGRGGRGGRGDG 97
D + N+ Q +R GSG G G GRG Y G G RG GG GG G G
Sbjct: 550 DLDGRNITVNQAQSR-------GSGGGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGGG 708
Query: 98 RGGFKRYGDDRTSHQDIARSNADGLYVG 125
GG YG+ R +R G Y G
Sbjct: 709 GGG--GYGERRGGGGGYSRGGGGGGYGG 786
>TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 754
Score = 41.2 bits (95), Expect = 5e-04
Identities = 32/88 (36%), Positives = 37/88 (41%), Gaps = 5/88 (5%)
Frame = +2
Query: 43 DAESDNVPRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGD-----RGRGGRGGRGDG 97
D + N+ Q +R GSG G G GRG Y G G RG GG GG G G
Sbjct: 296 DLDGRNITVNQAQSR-------GSGGGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGGG 454
Query: 98 RGGFKRYGDDRTSHQDIARSNADGLYVG 125
GG YG+ R +R G Y G
Sbjct: 455 GGG--GYGERRGGGGGYSRGGGGGGYGG 532
>BQ143793 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (19%)
Length = 1100
Score = 41.2 bits (95), Expect = 5e-04
Identities = 29/74 (39%), Positives = 36/74 (48%), Gaps = 12/74 (16%)
Frame = -3
Query: 62 DGDGSG---RGRGRGRGRDVYARGRGDRGRGGRG-GRGDG-------RGGFKRYGD-DRT 109
+G G G G GRG G+ + G G R RGG G GRG G +G KR+G DR
Sbjct: 312 EGKGEG*NNIGEGRGDGKRKWGGGGGGRRRGGEGMGRGGGG*MIFKEKGKGKRWGG*DRV 133
Query: 110 SHQDIARSNADGLY 123
++ R DG Y
Sbjct: 132 GEREGGRGRGDGCY 91
Score = 34.3 bits (77), Expect = 0.063
Identities = 20/49 (40%), Positives = 24/49 (48%), Gaps = 13/49 (26%)
Frame = -1
Query: 63 GDGSGRGR-------GRGRGRDVY------ARGRGDRGRGGRGGRGDGR 98
G G G G+ GRG+G+D GRG+ G GG GG G GR
Sbjct: 359 GGGGGGGKRKKKREGGRGKGKDEII*EKGGGMGRGNGGEGGGGGEGGGR 213
Score = 33.1 bits (74), Expect = 0.14
Identities = 26/88 (29%), Positives = 41/88 (46%)
Frame = -2
Query: 49 VPRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDR 108
V +R+ +++ + G+G G R +GRG G+G +G+ RG RG +GG +R
Sbjct: 550 VGKRKGESKWEQGQGEG-GDKRKKGRGE-----GKGGKGKTQRGKRGGKKGGRRR----- 404
Query: 109 TSHQDIARSNADGLYVGDNADGEKLAKK 136
++ G G GEK KK
Sbjct: 403 -GRREREMGYMKGSIRGGEGGGEKGKKK 323
Score = 32.3 bits (72), Expect = 0.24
Identities = 20/57 (35%), Positives = 27/57 (47%), Gaps = 17/57 (29%)
Frame = -2
Query: 64 DGSGRGRGRGRGRDVYARGRGDRGRGGRGGR-----------------GDGRGGFKR 103
+G GRG+GR RG+ + RG+G +G+ GR G G GG KR
Sbjct: 655 EGGGRGKGR-RGKRGWDRGKGSKGQETENGRRK*GKVGKRKGESKWEQGQGEGGDKR 488
Score = 30.4 bits (67), Expect = 0.91
Identities = 17/42 (40%), Positives = 24/42 (56%), Gaps = 3/42 (7%)
Frame = -2
Query: 67 GRGRGRGRGRD---VYARGRGDRGRGGRGGRGDGRGGFKRYG 105
GRG+G G G+ ++ G G G+GG+ RG +GG K G
Sbjct: 136 GRGKGGGEGKGGWMLWGMGGGGGGKGGKKRRGK-KGGKKERG 14
Score = 30.4 bits (67), Expect = 0.91
Identities = 17/37 (45%), Positives = 20/37 (53%)
Frame = -2
Query: 69 GRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYG 105
GRG+G G G+G G GG G G+GG KR G
Sbjct: 136 GRGKGGGE-----GKGGWMLWGMGGGGGGKGGKKRRG 41
Score = 30.4 bits (67), Expect = 0.91
Identities = 35/135 (25%), Positives = 56/135 (40%), Gaps = 3/135 (2%)
Frame = -3
Query: 9 GGGRGRGKPLEEAAQEAPQAPVVNRHIRVRQTPADAESDNVPRRQPMNRFVRDDG---DG 65
GGGRG G +V R +RV++ D ++ R+ R+ G
Sbjct: 630 GGGRGDG--------------IVVREVRVKRRKMDGANEE--------RWGRERGRVNGN 517
Query: 66 SGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSNADGLYVG 125
G+GRG R + + +RG+ G G+ +G + ++ + DI R G+ G
Sbjct: 516 RGKGRGGTSERKEEGKEKEERGKRKEGKGGERKG--EEGEEEGSGKWDI*R----GV*GG 355
Query: 126 DNADGEKLAKKLGPE 140
G+K KK G E
Sbjct: 354 GRGGGKKEKKKRGWE 310
>TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 912
Score = 41.2 bits (95), Expect = 5e-04
Identities = 32/88 (36%), Positives = 37/88 (41%), Gaps = 5/88 (5%)
Frame = +1
Query: 43 DAESDNVPRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGD-----RGRGGRGGRGDG 97
D + N+ Q +R GSG G G GRG Y G G RG GG GG G G
Sbjct: 454 DLDGRNITVNQAQSR-------GSGGGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGGG 612
Query: 98 RGGFKRYGDDRTSHQDIARSNADGLYVG 125
GG YG+ R +R G Y G
Sbjct: 613 GGG--GYGERRGGGGGYSRGGGGGGYGG 690
>TC85487 homologue to SP|P49688|RS2_ARATH 40S ribosomal protein S2.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (88%)
Length = 1114
Score = 40.4 bits (93), Expect = 9e-04
Identities = 33/103 (32%), Positives = 47/103 (45%), Gaps = 3/103 (2%)
Frame = +1
Query: 72 RGRGRDVYARGRGDRG--RGGRGGRGD-GRGGFKRYGDDRTSHQDIARSNADGLYVGDNA 128
RG R + G G RG RGGRGGRGD GRGG +R G R + G V
Sbjct: 82 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLV---- 249
Query: 129 DGEKLAKKLGPEIMDQITEAYEEIIERVLPSPLQDEYVEAMDI 171
E + L + + +II+ ++ L+DE ++ M +
Sbjct: 250 -KEGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPV 375
Score = 35.8 bits (81), Expect = 0.022
Identities = 24/45 (53%), Positives = 25/45 (55%), Gaps = 5/45 (11%)
Frame = +1
Query: 55 MNRFVRDDGDGSGRGRG-RGRGRDVYARG-RGDRGRGGR---GGR 94
+N GD G G G GRG D RG RGDRGRGGR GGR
Sbjct: 64 INNMAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGR 198
>TC76918 similar to GP|2231329|gb|AAB62000.1| bactinecin 11 {Ovis aries},
partial (13%)
Length = 576
Score = 40.4 bits (93), Expect = 9e-04
Identities = 29/64 (45%), Positives = 30/64 (46%), Gaps = 13/64 (20%)
Frame = +1
Query: 63 GDGSGRGRGRGRGR--------DVYARGRGDRGRG-----GRGGRGDGRGGFKRYGDDRT 109
G G GRGRGRGRG D Y GRG GRG GR RG GRG YG
Sbjct: 4 GRGRGRGRGRGRGMYNGGMEYGDGYDGGRGYGGRGRGRAWGRAFRGRGRG----YGAQPV 171
Query: 110 SHQD 113
+ D
Sbjct: 172 GYYD 183
Score = 33.1 bits (74), Expect = 0.14
Identities = 24/59 (40%), Positives = 25/59 (41%), Gaps = 20/59 (33%)
Frame = +1
Query: 61 DDGDG-SGRGRGRGRGRDVYARGRG-------------------DRGRGGRGGRGDGRG 99
D G G GRGRGR GR RGRG RG+G GRG GRG
Sbjct: 79 DGGRGYGGRGRGRAWGRAFRGRGRGYGAQPVGYYDNGEYDAPPAPRGQGRGRGRGRGRG 255
Score = 32.3 bits (72), Expect = 0.24
Identities = 13/15 (86%), Positives = 13/15 (86%)
Frame = +1
Query: 63 GDGSGRGRGRGRGRD 77
G G GRGRGRGRGRD
Sbjct: 217 GQGRGRGRGRGRGRD 261
Score = 28.1 bits (61), Expect = 4.5
Identities = 18/42 (42%), Positives = 22/42 (51%)
Frame = +1
Query: 53 QPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGR 94
QP+ + + D RG+GRG RGRG RGRG GR
Sbjct: 163 QPVGYYDNGEYDAPPAPRGQGRG-----RGRG-RGRGRDAGR 270
>TC85851 similar to PIR|T12180|T12180 probable transcription factor - fava
bean, partial (93%)
Length = 2007
Score = 39.3 bits (90), Expect = 0.002
Identities = 31/67 (46%), Positives = 36/67 (53%)
Frame = +3
Query: 41 PADAESDNVPRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
PA S P Q + R R++G GRG G GRG D R RG GRGG G G GRGG
Sbjct: 372 PALLPSKPTPPAQAV-RESRNEGGRGGRGFG-GRGGD---RDRGFGGRGGDRGFGGGRGG 536
Query: 101 FKRYGDD 107
+ +G D
Sbjct: 537 -RGFGRD 554
Score = 29.3 bits (64), Expect = 2.0
Identities = 14/26 (53%), Positives = 14/26 (53%)
Frame = +3
Query: 75 GRDVYARGRGDRGRGGRGGRGDGRGG 100
G Y G RGRGGRG RG G G
Sbjct: 1215 GESHYNSGGRGRGRGGRGARGGGFRG 1292
>TC79934 similar to GP|20259633|gb|AAM14173.1 putative GAR1 protein
{Arabidopsis thaliana}, partial (87%)
Length = 814
Score = 39.3 bits (90), Expect = 0.002
Identities = 22/47 (46%), Positives = 26/47 (54%)
Frame = +1
Query: 54 PMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
P+ RF+ + GRG GRG + RG G RG GG GRG RGG
Sbjct: 466 PLARFLPQPKGQASGGRGGGRGGGGFGRG-GGRGGGGFRGRGPPRGG 603
Score = 36.6 bits (83), Expect = 0.013
Identities = 22/39 (56%), Positives = 22/39 (56%)
Frame = +1
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGF 101
G G GRG G RGR GRG RGGRGG GRG F
Sbjct: 544 GRGGGRGGGGFRGRGPPRGGRGP-PRGGRGGGFRGRGRF 657
Score = 35.8 bits (81), Expect = 0.022
Identities = 22/37 (59%), Positives = 22/37 (59%), Gaps = 1/37 (2%)
Frame = +1
Query: 65 GSGRGRGRGRGRDVYARGRGDRGRG-GRGGRGDGRGG 100
G G GRG GRG G G RGRG RGGRG RGG
Sbjct: 532 GGGFGRGGGRG------GGGFRGRGPPRGGRGPPRGG 624
Score = 35.8 bits (81), Expect = 0.022
Identities = 26/58 (44%), Positives = 31/58 (52%), Gaps = 7/58 (12%)
Frame = +1
Query: 55 MNRFVRDDGDGSGRGRGRGRGRDVYARGRGD----RGRGG---RGGRGDGRGGFKRYG 105
+++ +R G GRG G GRD RG D RGRGG GGRG G GGF+ G
Sbjct: 40 LSQTMRPPARGGGRGGGFRGGRDGGFRGGRDGGGFRGRGGGRFGGGRGGG-GGFRDEG 210
Score = 28.9 bits (63), Expect = 2.6
Identities = 21/42 (50%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Frame = +1
Query: 68 RGRGRGRGRDVYARGRGDRGRGGRGGRG-DGRGGFKRYGDDR 108
RG GRG G + GR RGGR G G GRGG R+G R
Sbjct: 67 RGGGRGGG---FRGGRDGGFRGGRDGGGFRGRGG-GRFGGGR 180
Score = 27.7 bits (60), Expect = 5.9
Identities = 15/29 (51%), Positives = 18/29 (61%), Gaps = 1/29 (3%)
Frame = +1
Query: 81 RGRGDRGRGG-RGGRGDGRGGFKRYGDDR 108
+G+ GRGG RGG G GRGG + G R
Sbjct: 493 KGQASGGRGGGRGGGGFGRGGGRGGGGFR 579
>BQ137346 homologue to GP|15027095|em hypothetical protein LT.09 {Leishmania
major}, partial (0%)
Length = 1101
Score = 38.9 bits (89), Expect = 0.003
Identities = 34/114 (29%), Positives = 50/114 (43%), Gaps = 5/114 (4%)
Frame = +3
Query: 12 RGRGKPLEEAAQEAP--QAPVVNRHIR--VRQTPADAESDNVPRRQPMNRFVRDDGDGSG 67
R KP E A++A QAP N+ + PA +N P+ Q R
Sbjct: 216 RAGPKPKRENAKDAHKRQAPTTNKFSAGATQHPPARHTKNNTPKTQKQTTHDRQTTQNKS 395
Query: 68 RGRGRGRGRDVYARGRGDRGRGGR-GGRGDGRGGFKRYGDDRTSHQDIARSNAD 120
RG+ R ++ +GR DRG R G+G + G + G+DR Q +R A+
Sbjct: 396 RGK---RAKN---QGRPDRGTERRKAGQGGEKQGPAQQGNDRKKKQPASRGRAE 539
>TC77242 similar to GP|22655123|gb|AAM98152.1 putative protein {Arabidopsis
thaliana}, partial (43%)
Length = 1244
Score = 38.9 bits (89), Expect = 0.003
Identities = 26/51 (50%), Positives = 32/51 (61%), Gaps = 1/51 (1%)
Frame = +2
Query: 65 GSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYG-DDRTSHQDI 114
G RG G GRG+ + R RG RGRG G RG GRGG + G DD+ S +D+
Sbjct: 716 GEFRGPG-GRGQGI-RRNRG-RGRGSGGPRGGGRGGGRGRGRDDKVSAEDL 859
>TC85490 similar to GP|18146720|dbj|BAB82426. ribosomal protein S2
{Arabidopsis thaliana}, partial (89%)
Length = 1252
Score = 38.5 bits (88), Expect = 0.003
Identities = 31/102 (30%), Positives = 45/102 (43%)
Frame = +1
Query: 70 RGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSNADGLYVGDNAD 129
RG R + RG GDRGRGGRGG GRGG +R R + G V
Sbjct: 67 RGATGDRAGFGRGFGDRGRGGRGG-DRGRGGRRRGAGRREEEEKWVPVTKLGRLV----- 228
Query: 130 GEKLAKKLGPEIMDQITEAYEEIIERVLPSPLQDEYVEAMDI 171
E + L + + +II+ ++ L+DE ++ M +
Sbjct: 229 KEGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPV 354
Score = 33.1 bits (74), Expect = 0.14
Identities = 24/37 (64%), Positives = 25/37 (66%), Gaps = 1/37 (2%)
Frame = +1
Query: 63 GDGSGRGRGRG-RGRDVYARGRGDRGRGGRGGRGDGR 98
GD +G GRG G RGR RG GDRGRGGR RG GR
Sbjct: 79 GDRAGFGRGFGDRGRG--GRG-GDRGRGGR-RRGAGR 177
>BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox carteri f.
nagariensis}, partial (13%)
Length = 1223
Score = 38.1 bits (87), Expect = 0.004
Identities = 29/70 (41%), Positives = 34/70 (48%), Gaps = 3/70 (4%)
Frame = +2
Query: 65 GSGRGRGR--GRGRDVYARGRGDRGRGGRGGRGD-GRGGFKRYGDDRTSHQDIARSNADG 121
GS RGRGR G GR RGRG +G GG GRG RGG G + I N G
Sbjct: 224 GSERGRGRRGGGGRGGGRRGRGKKGGGGTKGRGKRERGG----GTEGGGKGKIEGRNGTG 391
Query: 122 LYVGDNADGE 131
G++ G+
Sbjct: 392 RRRGESGGGK 421
Score = 35.0 bits (79), Expect = 0.037
Identities = 31/104 (29%), Positives = 45/104 (42%), Gaps = 3/104 (2%)
Frame = +2
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGG--RGDG-RGGFKRYGDDRTSHQDIARSNA 119
G+G G G G + RGRG RG GGRGG RG G +GG G +
Sbjct: 188 GEGGEGGGGAEGGSE---RGRGRRGGGGRGGGRRGRGKKGGGGTKGRGKRERGGGTEGGG 358
Query: 120 DGLYVGDNADGEKLAKKLGPEIMDQITEAYEEIIERVLPSPLQD 163
G G N G + + G + ++ E+++ + S LQ+
Sbjct: 359 KGKIEGRNGTGRRRGESGGGKGREK-----SELLQTDVESSLQE 475
Score = 34.7 bits (78), Expect = 0.048
Identities = 18/44 (40%), Positives = 24/44 (53%)
Frame = +3
Query: 65 GSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDR 108
GSG GRG G R + G GG GG G G GG ++ G+++
Sbjct: 180 GSGGRGGRGGGERRGGRRGEEGGGGGGGGEGGGGGGGRKGGEEQ 311
Score = 32.3 bits (72), Expect = 0.24
Identities = 20/41 (48%), Positives = 23/41 (55%), Gaps = 1/41 (2%)
Frame = +2
Query: 60 RDDGDGSGRGR-GRGRGRDVYARGRGDRGRGGRGGRGDGRG 99
R G G G GR GRG+ +GRG R RGG G G G+G
Sbjct: 245 RRGGGGRGGGRRGRGKKGGGGTKGRGKRERGG-GTEGGGKG 364
Score = 32.0 bits (71), Expect = 0.31
Identities = 18/38 (47%), Positives = 22/38 (57%), Gaps = 2/38 (5%)
Frame = +2
Query: 68 RGRGRGRGRDVYARGRGDRGRGGRGGRGDG--RGGFKR 103
RGRG G+ R RG+RG+ G GG G G GG +R
Sbjct: 137 RGRGSGK-----MRRRGERGKRGEGGEGGGGAEGGSER 235
Score = 32.0 bits (71), Expect = 0.31
Identities = 25/57 (43%), Positives = 29/57 (50%), Gaps = 10/57 (17%)
Frame = +2
Query: 63 GDGSGRGRGRG-RGRDVY-------ARGRGDRGRG--GRGGRGDGRGGFKRYGDDRT 109
G GSG+ R RG RG+ A G +RGRG G GGRG GR G + G T
Sbjct: 140 GRGSGKMRRRGERGKRGEGGEGGGGAEGGSERGRGRRGGGGRGGGRRGRGKKGGGGT 310
Score = 27.3 bits (59), Expect = 7.7
Identities = 18/52 (34%), Positives = 24/52 (45%)
Frame = +1
Query: 81 RGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSNADGLYVGDNADGEK 132
RGR R G GGR GRG G+ R + + RS + + A GE+
Sbjct: 859 RGRETRSETGGGGRERGRGD----GERRRRERGMTRSRVERITEV*GARGER 1002
>TC87186 similar to PIR|F96687|F96687 hypothetical protein T6J19.1
[imported] - Arabidopsis thaliana, partial (47%)
Length = 1894
Score = 37.4 bits (85), Expect = 0.007
Identities = 18/33 (54%), Positives = 21/33 (63%)
Frame = +1
Query: 66 SGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGR 98
S RGRGRGRG+ + RG G G G R G +GR
Sbjct: 325 SNRGRGRGRGKALSGRGSGTLGGGRRTGAVNGR 423
Score = 31.2 bits (69), Expect = 0.53
Identities = 15/31 (48%), Positives = 16/31 (51%)
Frame = +3
Query: 65 GSGRGRGRGRGRDVYARGRGDRGRGGRGGRG 95
G G RG + RGRG GRGG GRG
Sbjct: 981 GGXXGGSRGGSGSMRGRGRGRPGRGGGXGRG 1073
>TC92013 homologue to GP|11595557|emb|CAC18142. related to c-module-binding
factor {Neurospora crassa}, partial (2%)
Length = 1437
Score = 37.4 bits (85), Expect = 0.007
Identities = 33/100 (33%), Positives = 39/100 (39%), Gaps = 4/100 (4%)
Frame = -2
Query: 4 PKVLPGGGRGRGKPLEEAAQEAPQAPVVNR----HIRVRQTPADAESDNVPRRQPMNRFV 59
P P RG+P P P V R + + +ES++ P R
Sbjct: 434 PPPPPTSSSRRGRP--------PAGPTVVRLGESDLESESEISSSESESGIGGSPAMRGP 279
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRG 99
G G GRGRG GRGR GRG GRG GRG
Sbjct: 278 SMRGRGGGRGRGGGRGR----------GRGRGRGRGRGRG 189
Score = 33.9 bits (76), Expect = 0.082
Identities = 17/26 (65%), Positives = 18/26 (68%)
Frame = -2
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGD 85
R G G GRGRGRGRGR RGRG+
Sbjct: 254 RGRGGGRGRGRGRGRGR---GRGRGN 186
Score = 30.0 bits (66), Expect = 1.2
Identities = 17/36 (47%), Positives = 18/36 (49%)
Frame = -2
Query: 73 GRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDR 108
G G RG RGRGG GRG GRG + G R
Sbjct: 311 GIGGSPAMRGPSMRGRGGGRGRGGGRGRGRGRGRGR 204
Score = 29.3 bits (64), Expect = 2.0
Identities = 14/23 (60%), Positives = 14/23 (60%)
Frame = -2
Query: 60 RDDGDGSGRGRGRGRGRDVYARG 82
R G G GRGRGRGRG A G
Sbjct: 236 RGRGRGRGRGRGRGRGN*SVAEG 168
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.137 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,283,599
Number of Sequences: 36976
Number of extensions: 84651
Number of successful extensions: 1738
Number of sequences better than 10.0: 200
Number of HSP's better than 10.0 without gapping: 906
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1324
length of query: 310
length of database: 9,014,727
effective HSP length: 96
effective length of query: 214
effective length of database: 5,465,031
effective search space: 1169516634
effective search space used: 1169516634
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC147483.4


