

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
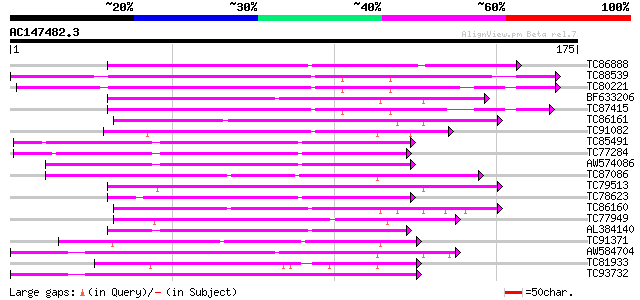
Query= AC147482.3 - phase: 0
(175 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86888 similar to PIR|S71274|S71274 blue copper-binding protein... 106 5e-24
TC88539 weakly similar to GP|9294157|dbj|BAB02059.1 blue copper-... 75 1e-14
TC80221 weakly similar to GP|9294157|dbj|BAB02059.1 blue copper-... 72 9e-14
BF633206 similar to GP|21553614|gb| blue copper protein putativ... 66 6e-12
TC87415 weakly similar to GP|9294157|dbj|BAB02059.1 blue copper-... 64 4e-11
TC86161 weakly similar to PIR|T01605|T01605 phytocyanin At2g4479... 62 9e-11
TC91082 similar to PIR|T01852|T01852 probable blue copper-bindin... 61 3e-10
TC85491 homologue to GP|6688810|emb|CAB65280.1 basic blue protei... 59 8e-10
TC77284 weakly similar to GP|6688810|emb|CAB65280.1 basic blue p... 59 1e-09
AW574086 homologue to GP|6688810|emb basic blue protein {Medicag... 59 1e-09
TC87086 similar to SP|Q41001|BCP_PEA Blue copper protein precurs... 58 2e-09
TC79513 weakly similar to GP|9885806|gb|AAG01535.1| stellacyanin... 58 2e-09
TC78623 similar to GP|3860333|emb|CAA10134.1 basic blue copper p... 58 2e-09
TC86160 weakly similar to PIR|T01605|T01605 phytocyanin At2g4479... 57 3e-09
TC77949 weakly similar to GP|11762218|gb|AAG40387.1 AT4g27520 {A... 55 1e-08
AL384140 weakly similar to GP|2094888|pdb|2 Cucumber Basic Prote... 54 3e-08
TC91371 weakly similar to GP|14140127|emb|CAC39044. uclacyanin 3... 52 1e-07
AW584704 similar to SP|P05790|FBOH Fibroin heavy chain precursor... 51 2e-07
TC81933 similar to SP|P10496|GRP2_PHAVU Glycine-rich cell wall s... 51 2e-07
TC93732 homologue to OMNI|NT01MC2635 Integral membrane protein o... 51 2e-07
>TC86888 similar to PIR|S71274|S71274 blue copper-binding protein 15K -
Arabidopsis thaliana, partial (14%)
Length = 697
Score = 106 bits (264), Expect = 5e-24
Identities = 52/128 (40%), Positives = 74/128 (57%)
Frame = +1
Query: 31 HKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTFIRN 90
H VG ++GW +NYT W++ +YVGD++ F + K YNV VN+TGYD C + + N
Sbjct: 82 HIVGANRGWNPGINYTLWANNHTIYVGDYISFRYQKNQYNVFLVNQTGYDNCTLDSAVGN 261
Query: 91 LTRGGRDVVQLTEAKTYYFITGGGYCFHGMKVAVDVQEHPTPAPSPSLSDTAKSGGDSIL 150
+ G+D + ++ YYFI G G C +GMKV+V V HP P+P PS S S +S
Sbjct: 262 WS-SGKDFILFNKSMRYYFICGNGQCNNGMKVSVFV--HPLPSPPPSSSQHNHSSPNSAA 432
Query: 151 PSMYTCFG 158
P + G
Sbjct: 433 PMVLEYLG 456
>TC88539 weakly similar to GP|9294157|dbj|BAB02059.1 blue copper-binding
protein-like {Arabidopsis thaliana}, partial (37%)
Length = 835
Score = 75.1 bits (183), Expect = 1e-14
Identities = 54/172 (31%), Positives = 88/172 (50%), Gaps = 2/172 (1%)
Frame = +3
Query: 1 MKNSRVLLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWL 60
M +SRV+L++S+ + +LS + H VG KGW + +YT W+ + VGD L
Sbjct: 105 MASSRVVLILSISMVLLSSVAIAATD----HIVGDDKGWTVDFDYTQWAQDKVFRVGDNL 272
Query: 61 KFVFDKRYYNVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQL-TEAKTYYFITGGGYC-FH 118
F +D +NV +VN T + C L+ G+D++QL TE + +Y +C
Sbjct: 273 VFNYDPARHNVFKVNGTLFQSCTFPPKNEALST-GKDIIQLKTEGRKWYVCGVADHCSAR 449
Query: 119 GMKVAVDVQEHPTPAPSPSLSDTAKSGGDSILPSMYTCFGIIVANVVYVSLV 170
MK+ + V PAPSP S A S S+ FG+++A +V ++++
Sbjct: 450 QMKLVITVLAEGAPAPSPPPSSDAHSVVSSL-------FGVVMAIMVAIAVI 584
>TC80221 weakly similar to GP|9294157|dbj|BAB02059.1 blue copper-binding
protein-like {Arabidopsis thaliana}, partial (40%)
Length = 813
Score = 72.4 bits (176), Expect = 9e-14
Identities = 54/170 (31%), Positives = 82/170 (47%), Gaps = 2/170 (1%)
Frame = +1
Query: 3 NSRVLLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLKF 62
N L V+++ + L V M H VG KGW + NYT W+ + VGD L F
Sbjct: 61 NQMALNRVAILAISMVLLSSVAMAAD--HIVGDDKGWTVDFNYTQWTQDKVFRVGDNLVF 234
Query: 63 VFDKRYYNVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQL-TEAKTYYFITGGGYC-FHGM 120
+D +N+ +VN T + C L+ G+D++QL TE + +Y +C H M
Sbjct: 235 NYDNTKHNIFKVNGTLFKDCTFPPKNEALST-GKDIIQLKTEGRKWYVCGVADHCSAHQM 411
Query: 121 KVAVDVQEHPTPAPSPSLSDTAKSGGDSILPSMYTCFGIIVANVVYVSLV 170
K + V PAPSP S S SI+ SM FG+++ +V ++ +
Sbjct: 412 KFVITVLAEGAPAPSPPPS----SNAHSIVSSM---FGVVMVAIVAMATI 540
>BF633206 similar to GP|21553614|gb| blue copper protein putative
{Arabidopsis thaliana}, partial (56%)
Length = 478
Score = 66.2 bits (160), Expect = 6e-12
Identities = 43/122 (35%), Positives = 64/122 (52%), Gaps = 4/122 (3%)
Frame = +2
Query: 31 HKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTFIRN 90
H VGGS+GW + ++ +WSS + VGD L F + + V +++ Y C D++ N
Sbjct: 113 HNVGGSQGWDPSSDFDSWSSGQTFKVGDQLVFKYTSMHSVVELSDESAYKKC-DISTPLN 289
Query: 91 LTRGGRDVVQLTEAKTYYFITGG-GYCFHGMKVAVDV---QEHPTPAPSPSLSDTAKSGG 146
G+DVV+L + T YF G G+C GMKV + V + A SPS S ++ S
Sbjct: 290 SLSTGKDVVKLDKPGTRYFTCGTLGHCDQGMKVKITVGNGNGSSSTASSPSSSSSSPSSS 469
Query: 147 DS 148
S
Sbjct: 470 SS 475
>TC87415 weakly similar to GP|9294157|dbj|BAB02059.1 blue copper-binding
protein-like {Arabidopsis thaliana}, partial (31%)
Length = 710
Score = 63.5 bits (153), Expect = 4e-11
Identities = 44/140 (31%), Positives = 70/140 (49%), Gaps = 2/140 (1%)
Frame = +1
Query: 31 HKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTFIRN 90
H VG KGW + NYT W+ + VGD L F +D +NV +V+ + C +
Sbjct: 139 HVVGDEKGWTVDFNYTQWAQDKVFRVGDNLVFNYDNTKHNVFKVDGKLFQSCTFPSENEA 318
Query: 91 LTRGGRDVVQL-TEAKTYYFITGGGYC-FHGMKVAVDVQEHPTPAPSPSLSDTAKSGGDS 148
L+ G+DV+QL TE + +Y +C MK+ ++V E P+PS S S
Sbjct: 319 LST-GKDVIQLKTEGRKWYVCGKANHCAARQMKLVINVLEEGAPSPS--------SSAHS 471
Query: 149 ILPSMYTCFGIIVANVVYVS 168
I+ S+ FG+I+ + ++
Sbjct: 472 IVSSI---FGVIMVATIAIA 522
>TC86161 weakly similar to PIR|T01605|T01605 phytocyanin At2g44790
[imported] - Arabidopsis thaliana, partial (17%)
Length = 736
Score = 62.4 bits (150), Expect = 9e-11
Identities = 42/131 (32%), Positives = 62/131 (47%), Gaps = 11/131 (8%)
Frame = +1
Query: 33 VGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTFIRNLT 92
VG + GW + V+YT W+S + VGD L F + ++ V EV+++GY C I++
Sbjct: 175 VGDANGWTQGVDYTKWASGKTFKVGDNLVFKYGS-FHQVNEVDESGYKSCSTSNTIKSYD 351
Query: 93 RGGRDVVQLTEAKTYYFITGGGYCFH--GMKVAVDV---------QEHPTPAPSPSLSDT 141
G V K Y+ G+C GMK+ V+V P P SPS + +
Sbjct: 352 DGDSKVPLTKAGKIYFICPTPGHCTSTGGMKLEVNVVAASTTPTPSGTPPPTKSPSTTPS 531
Query: 142 AKSGGDSILPS 152
A S +S PS
Sbjct: 532 APSETNSTTPS 564
>TC91082 similar to PIR|T01852|T01852 probable blue copper-binding protein
F9D12.16 - Arabidopsis thaliana, partial (55%)
Length = 684
Score = 60.8 bits (146), Expect = 3e-10
Identities = 36/116 (31%), Positives = 59/116 (50%), Gaps = 8/116 (6%)
Frame = +2
Query: 30 LHKVGGSKGWKE--NVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTF 87
++KVG S GW N++Y W++ ++ +GD + F + +++NV+ V Y C +
Sbjct: 26 VYKVGDSAGWTTLGNIDYKKWAATKNFQLGDTIIFEYSAKFHNVMRVTHAMYKSCNASSP 205
Query: 88 IRNLTRGGRDVVQLTEAKTYYFITG-GGYCFHGMKV-----AVDVQEHPTPAPSPS 137
I T G D +++T ++F G G+C G KV V V P P+ SPS
Sbjct: 206 IATFTT-GNDTIKITNHGHHFFFCGVPGHCQAGQKVDINVLKVSVAASPAPSSSPS 370
>TC85491 homologue to GP|6688810|emb|CAB65280.1 basic blue protein {Medicago
sativa subsp. x varia}, complete
Length = 752
Score = 59.3 bits (142), Expect = 8e-10
Identities = 35/124 (28%), Positives = 63/124 (50%)
Frame = +3
Query: 2 KNSRVLLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLK 61
+ S + MV +V+ +L L + + + VGG KGW + W + + GD L
Sbjct: 84 RGSAAMNMV-IVISLLCLMVLAKSTNAETYTVGGPKGWTFGIK--KWPNGKSFVAGDVLD 254
Query: 62 FVFDKRYYNVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITGGGYCFHGMK 121
F ++ + +NV+ V++TGYD C + + R G D ++L + Y+ G+C GMK
Sbjct: 255 FGYNPKMHNVVLVDQTGYDKCKTPEGSK-VFRTGSDQIELVKGDNYFICNLPGHCQSGMK 431
Query: 122 VAVD 125
+ ++
Sbjct: 432 IYIN 443
>TC77284 weakly similar to GP|6688810|emb|CAB65280.1 basic blue protein
{Medicago sativa subsp. x varia}, complete
Length = 814
Score = 58.9 bits (141), Expect = 1e-09
Identities = 35/123 (28%), Positives = 64/123 (51%)
Frame = +2
Query: 2 KNSRVLLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLK 61
+ S + MV+++ +L L + + VGG+ GW N + TW + + GD L
Sbjct: 104 RGSASMNMVTLI-SLLCLLVLAESANAASYTVGGTGGWTYNTD--TWPNGKKFKAGDVLS 274
Query: 62 FVFDKRYYNVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITGGGYCFHGMK 121
F +D +NV+ V+K+GY+ C + + G D ++L+ + Y+ + G+C GMK
Sbjct: 275 FNYDSTTHNVVAVDKSGYNNCKTPGGAK-VFSSGSDQIRLSRGQNYFICSYPGHCQSGMK 451
Query: 122 VAV 124
V++
Sbjct: 452 VSI 460
>AW574086 homologue to GP|6688810|emb basic blue protein {Medicago sativa
subsp. x varia}, partial (97%)
Length = 504
Score = 58.9 bits (141), Expect = 1e-09
Identities = 32/114 (28%), Positives = 58/114 (50%)
Frame = +1
Query: 12 VVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNV 71
+V+ +L L + + + VGG KGW + W + + GD L F ++ + +NV
Sbjct: 4 IVISLLCLMVLAKSTNAETYTVGGPKGWTFGIK--KWPNGKSFVAGDVLDFGYNPKMHNV 177
Query: 72 LEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITGGGYCFHGMKVAVD 125
+ V++TGYD C + + R G D ++L + Y+ G+C GMK+ ++
Sbjct: 178 VLVDQTGYDKCKTPEGSK-VFRTGSDQIELVKGDNYFICNLPGHCQSGMKIYIN 336
>TC87086 similar to SP|Q41001|BCP_PEA Blue copper protein precursor. [Garden
pea] {Pisum sativum}, partial (87%)
Length = 1219
Score = 58.2 bits (139), Expect = 2e-09
Identities = 40/136 (29%), Positives = 65/136 (47%), Gaps = 1/136 (0%)
Frame = +1
Query: 12 VVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNV 71
++LG + + V +H VG GW +Y TW+S + VGD L F + + V
Sbjct: 451 LILGFFLVINVAVPTLATVHTVGDKSGWAIGSDYNTWASDKTFAVGDSLVFNYGAG-HTV 627
Query: 72 LEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITG-GGYCFHGMKVAVDVQEHP 130
EV ++ Y C I + G + L +A T+YFI G+C GMK++V V+
Sbjct: 628 DEVKESDYKSCTTGNSI-STDSSGPTTIPLKKAGTHYFICAVPGHCTGGMKLSVKVKASS 804
Query: 131 TPAPSPSLSDTAKSGG 146
+ + +PS + + G
Sbjct: 805 SASSAPSATPSPSGKG 852
>TC79513 weakly similar to GP|9885806|gb|AAG01535.1| stellacyanin-like
protein CASLP1 precursor {Capsicum annuum}, partial
(33%)
Length = 1093
Score = 58.2 bits (139), Expect = 2e-09
Identities = 37/129 (28%), Positives = 57/129 (43%), Gaps = 7/129 (5%)
Frame = +3
Query: 31 HKVGGSKGWKENVN----YTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMT 86
H VG + GW N YT W+S + VGD L F + ++V +V KT YD C
Sbjct: 477 HVVGDTTGWTIPTNGASFYTNWASNKTFTVGDTLVFNYASGQHDVAKVTKTAYDSCNGAN 656
Query: 87 FIRNLTRGGRDVVQLTEAKTYYFITGGGYCFHGMKVAVDV---QEHPTPAPSPSLSDTAK 143
+ LT V + + G+C G K++++V P AP+PS S
Sbjct: 657 TLFTLTNSPATVTLNETGQQNFICAVPGHCSAGQKLSINVVKASASPVSAPTPSASPPKA 836
Query: 144 SGGDSILPS 152
+ + +P+
Sbjct: 837 TPAPTPVPA 863
>TC78623 similar to GP|3860333|emb|CAA10134.1 basic blue copper protein
{Cicer arietinum}, partial (98%)
Length = 743
Score = 57.8 bits (138), Expect = 2e-09
Identities = 31/95 (32%), Positives = 48/95 (49%)
Frame = +3
Query: 31 HKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTFIRN 90
+ VGG GW N W + + GD L F + +NV+ VNK GYD C +
Sbjct: 114 YTVGGPGGW--TFNTVGWPNGKRFRAGDTLVFNYSPSAHNVVAVNKGGYDSCKTPRGAK- 284
Query: 91 LTRGGRDVVQLTEAKTYYFITGGGYCFHGMKVAVD 125
+ R G+D ++L + Y+ G+C GMK+A++
Sbjct: 285 VYRSGKDQIRLARGQNYFICNFVGHCESGMKIAIN 389
>TC86160 weakly similar to PIR|T01605|T01605 phytocyanin At2g44790
[imported] - Arabidopsis thaliana, partial (34%)
Length = 918
Score = 57.4 bits (137), Expect = 3e-09
Identities = 48/129 (37%), Positives = 64/129 (49%), Gaps = 9/129 (6%)
Frame = +3
Query: 33 VGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTFIRNLT 92
VG + GW V+YT W+S + VGD L F + + V EV+++ Y C I+N
Sbjct: 195 VGDANGWNLGVDYTKWASGKTFKVGDNLVFKYGSS-HQVDEVDESDYKSCTSSNAIKNYA 371
Query: 93 RGGRDVVQLTEAKTYYFITGG-GYCFH--GMKVAVDV---QEHPTPA--PSPSLS-DTAK 143
GG V LT+A YFI G+C GMK+ V+V PTP+ P P+ S T
Sbjct: 372 -GGNSKVPLTKAGKIYFICPTLGHCTSTGGMKLEVNVVAASTTPTPSGTPPPTKSPSTTP 548
Query: 144 SGGDSILPS 152
S S PS
Sbjct: 549 STTPSTTPS 575
>TC77949 weakly similar to GP|11762218|gb|AAG40387.1 AT4g27520 {Arabidopsis
thaliana}, partial (34%)
Length = 1297
Score = 55.1 bits (131), Expect = 1e-08
Identities = 37/110 (33%), Positives = 51/110 (45%), Gaps = 3/110 (2%)
Frame = +3
Query: 33 VGGSKGWKENV--NYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTFIRN 90
VGG GW N NY W+ + + D + F + K +VLEV K Y+ C I+
Sbjct: 195 VGGKDGWTLNPSENYNQWAGRNRFQISDTIVFKYKKGSDSVLEVKKEDYEKCNKTNPIKK 374
Query: 91 LTRGGRDVVQLTEAKTYYFITGGGY-CFHGMKVAVDVQEHPTPAPSPSLS 139
G + L A +YFI+G C +G K+ + V TP SPS S
Sbjct: 375 FEDGETEFT-LDRAGPFYFISGKDQNCENGQKLTLVVISPRTPKSSPSPS 521
>AL384140 weakly similar to GP|2094888|pdb|2 Cucumber Basic Protein A Blue
Copper Protein, partial (92%)
Length = 441
Score = 53.9 bits (128), Expect = 3e-08
Identities = 32/94 (34%), Positives = 45/94 (47%)
Frame = +2
Query: 31 HKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTFIRN 90
H VG KGW V W + + GD L F + +NV+ VNK+GYD C+ +
Sbjct: 131 HIVGDGKGWSFGVQ--NWPAGKTFKAGDTLVFNYSPTSHNVVVVNKSGYDSCVAPKGSKV 304
Query: 91 LTRGGRDVVQLTEAKTYYFITGGGYCFHGMKVAV 124
T G D + L + Y+ G+C G K+AV
Sbjct: 305 YT-SGADRITLAKGGNYFLCGFPGHCNLGQKIAV 403
>TC91371 weakly similar to GP|14140127|emb|CAC39044. uclacyanin 3-like
protein {Oryza sativa}, partial (48%)
Length = 686
Score = 52.0 bits (123), Expect = 1e-07
Identities = 39/116 (33%), Positives = 58/116 (49%), Gaps = 4/116 (3%)
Frame = +1
Query: 16 VLSLWPMVVMGGPKL---HKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVL 72
VL + + M P L + VG S GW+ V Y+ W+S++ VGD L F++ + V
Sbjct: 64 VLCFFLAINMALPTLATFYTVGDSLGWQIGVEYSKWTSEKTFVVGDSLVFLYG-AIHTVD 240
Query: 73 EVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITGG-GYCFHGMKVAVDVQ 127
EV + Y C I + G + L A T+YFI+ G C GM++AV V+
Sbjct: 241 EVAASDYISCTTGNPISS-DNSGETTIALKTAGTHYFISATFGDCSSGMRLAVKVE 405
>AW584704 similar to SP|P05790|FBOH Fibroin heavy chain precursor (Fib-H)
(H-fibroin). [Silk moth] {Bombyx mori}, partial (0%)
Length = 679
Score = 51.2 bits (121), Expect = 2e-07
Identities = 41/144 (28%), Positives = 68/144 (46%), Gaps = 5/144 (3%)
Frame = +1
Query: 1 MKNSRVLLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWL 60
M SR L + +++ + S M K VG GW V+Y W++ + +GD L
Sbjct: 52 MALSRSLFLFALIATIFS-----TMAVAKDFVVGDESGWTLGVDYQAWAANKVFRLGDTL 216
Query: 61 KFVFDKRYYNVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITG-GGYCFHG 119
F + NV+ VN + + C + + + G D + LT ++I+G +C +G
Sbjct: 217 TFKYVAWKDNVVRVNGSDFQSC-SVPWAAPVLTSGHDKIALTTYGRRWYISGVANHCENG 393
Query: 120 MKVAVDV---QEHPTPAP-SPSLS 139
K+ ++V Q+ PAP SPS S
Sbjct: 394 QKLFINVLPKQDGWYPAPSSPSAS 465
>TC81933 similar to SP|P10496|GRP2_PHAVU Glycine-rich cell wall structural
protein 1.8 precursor (GRP 1.8). [Kidney bean French
bean], partial (4%)
Length = 765
Score = 51.2 bits (121), Expect = 2e-07
Identities = 42/124 (33%), Positives = 56/124 (44%), Gaps = 23/124 (18%)
Frame = +2
Query: 27 GPKLHKVGGSKGWKEN-------VNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGY 79
G K H VGG+ GW N NY++W+S + +GD+L F + +L NKT Y
Sbjct: 68 GYKNHTVGGTSGWLFNSSTNTSATNYSSWASSQSFNLGDYLIFNTNSNQSVILTYNKTAY 247
Query: 80 DYCI----DM-TFIRNLTRGGRD--------VVQLTEAKTYYFITG---GGYCFHGMKVA 123
C D+ TFI N GG + V LT YF + G C HG+
Sbjct: 248 TSCTADDSDVGTFIYN---GGTNNFSETLTIPVPLTIVGPNYFFSDTNEGVQCQHGLAFQ 418
Query: 124 VDVQ 127
+DVQ
Sbjct: 419 IDVQ 430
>TC93732 homologue to OMNI|NT01MC2635 Integral membrane protein of unknown
function {Magnetococcus sp. MC-1}, partial (2%)
Length = 443
Score = 51.2 bits (121), Expect = 2e-07
Identities = 34/127 (26%), Positives = 57/127 (44%)
Frame = +3
Query: 1 MKNSRVLLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWL 60
M SR L + +++ + S M K VG KGW +Y TW++ + +GD L
Sbjct: 54 MALSRALFLFALIASIFS-----TMAVAKDFVVGDEKGWTTLFDYQTWTANKVFRLGDTL 218
Query: 61 KFVFDKRYYNVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITGGGYCFHGM 120
F + NV+ VN + + C LT G ++ T + +Y + +C +G
Sbjct: 219 TFNYVGGKDNVVRVNGSDFKSCSVPLTAPVLTSGQDKIIITTYGRRWYISSVTDHCENGQ 398
Query: 121 KVAVDVQ 127
K+ + VQ
Sbjct: 399 KLFITVQ 419
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.140 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,553,377
Number of Sequences: 36976
Number of extensions: 103154
Number of successful extensions: 564
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 537
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 547
length of query: 175
length of database: 9,014,727
effective HSP length: 90
effective length of query: 85
effective length of database: 5,686,887
effective search space: 483385395
effective search space used: 483385395
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC147482.3


