

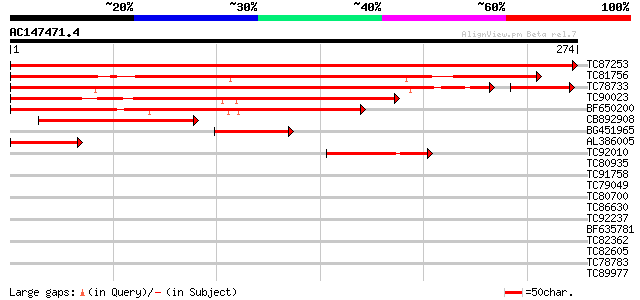
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147471.4 + phase: 0
(274 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87253 similar to GP|21592813|gb|AAM64762.1 ids4-like protein {... 542 e-155
TC81756 similar to GP|16506648|gb|AAL17697.1 IDS-4-like protein ... 292 8e-80
TC78733 similar to GP|21592813|gb|AAM64762.1 ids4-like protein {... 256 3e-75
TC90023 similar to PIR|G84886|G84886 hypothetical protein At2g45... 187 4e-48
BF650200 similar to GP|15215822|gb AT5g15330/F8M21_220 {Arabidop... 150 7e-37
CB892908 133 8e-32
BG451965 similar to GP|16506648|gb| IDS-4-like protein {Castanea... 80 1e-15
AL386005 weakly similar to PIR|G84886|G848 hypothetical protein ... 45 3e-05
TC92010 41 6e-04
TC80935 similar to GP|11994328|dbj|BAB02287. receptor protein-li... 38 0.005
TC91758 similar to GP|11994328|dbj|BAB02287. receptor protein-li... 34 0.053
TC79049 weakly similar to PIR|T10798|T10798 pherophorin-S - Volv... 31 0.45
TC80700 similar to GP|9759160|dbj|BAB09716.1 contains similarity... 30 1.3
TC86630 similar to GP|22136930|gb|AAM91809.1 unknown protein {Ar... 29 1.7
TC92237 similar to PIR|B96712|B96712 probable receptor protein F... 29 2.2
BF635781 similar to GP|17104621|gb| unknown protein {Arabidopsis... 28 2.9
TC82362 similar to GP|15450519|gb|AAK96552.1 AT4g22540/F7K2_120 ... 28 3.8
TC82605 similar to GP|17104721|gb|AAL34249.1 unknown protein {Ar... 28 4.9
TC78783 similar to GP|21537171|gb|AAM61512.1 unknown {Arabidopsi... 27 6.5
TC89977 similar to GP|5764395|gb|AAD51282.1| far-red impaired re... 27 6.5
>TC87253 similar to GP|21592813|gb|AAM64762.1 ids4-like protein {Arabidopsis
thaliana}, partial (80%)
Length = 1285
Score = 542 bits (1396), Expect = e-155
Identities = 274/274 (100%), Positives = 274/274 (100%)
Frame = +2
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEV 60
MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEV
Sbjct: 80 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEV 259
Query: 61 TKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAKSSDIELMTVGREIVDF 120
TKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAKSSDIELMTVGREIVDF
Sbjct: 260 TKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAKSSDIELMTVGREIVDF 439
Query: 121 HGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNKLVKECE 180
HGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNKLVKECE
Sbjct: 440 HGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNKLVKECE 619
Query: 181 VMLSIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQNMENIFIKL 240
VMLSIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQNMENIFIKL
Sbjct: 620 VMLSIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQNMENIFIKL 799
Query: 241 TTSALDTLKEIRGGSSTVSIYSLPPLHSETLVED 274
TTSALDTLKEIRGGSSTVSIYSLPPLHSETLVED
Sbjct: 800 TTSALDTLKEIRGGSSTVSIYSLPPLHSETLVED 901
>TC81756 similar to GP|16506648|gb|AAL17697.1 IDS-4-like protein {Castanea
sativa}, partial (68%)
Length = 1061
Score = 292 bits (748), Expect = 8e-80
Identities = 164/259 (63%), Positives = 191/259 (73%), Gaps = 2/259 (0%)
Frame = +3
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEV 60
MKFWKIL +QIEQTLP+WRD+FLSYK+LKKQLK + PK+ + + D
Sbjct: 39 MKFWKILKSQIEQTLPEWRDQFLSYKNLKKQLKAMCPKDAHTP-----PIWD-------- 179
Query: 61 TKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAKS-SDIELMTVGREIVD 119
+V FL LLEVEI+KFN FFV KEEEY+IKWKELQD+V SD+ELM++ REIVD
Sbjct: 180 AHQVSHFLCLLEVEIDKFNTFFVNKEEEYIIKWKELQDRVDRVMDYSDLELMSLWREIVD 359
Query: 120 FHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNKLVKEC 179
FHGEMVLLENYSALNYTGLVKIIKK+DKRTG LLRLPFIQ+VLNQPFF+ DVLN LVKEC
Sbjct: 360 FHGEMVLLENYSALNYTGLVKIIKKHDKRTGGLLRLPFIQEVLNQPFFETDVLNNLVKEC 539
Query: 180 EVMLSIIFPKS-GPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQNMENIFI 238
EV+L+I+F + P STSE E E KE L VP+E +EI+NMEN+ I
Sbjct: 540 EVLLNILFTNNDSPSCPCTSTSEENE----------ENKEKLMQVPEEIAEIENMENVLI 689
Query: 239 KLTTSALDTLKEIRGGSST 257
KLT SAL TL+EIRG SST
Sbjct: 690 KLTLSALRTLEEIRGQSST 746
>TC78733 similar to GP|21592813|gb|AAM64762.1 ids4-like protein {Arabidopsis
thaliana}, partial (83%)
Length = 1260
Score = 256 bits (654), Expect(2) = 3e-75
Identities = 139/238 (58%), Positives = 171/238 (71%), Gaps = 4/238 (1%)
Frame = +1
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEI--DSSCSKRRRLDDDGGAEG 58
MKF K L++QIE+TLP+WRDKFLSYK+LKK+LK + P D KR ++D G
Sbjct: 217 MKFGKSLSSQIEKTLPEWRDKFLSYKELKKKLKSLEPASASADDRPVKRLKVDSGNADAG 396
Query: 59 EVTKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAKSSDIELMTVGREIV 118
E++KE DF LLE E+EKFN FFVEKEEEY+I+ KELQD+VA K E+M + +EIV
Sbjct: 397 EMSKEESDFRNLLENELEKFNNFFVEKEEEYIIRLKELQDRVAKVKDYSEEMMKIRKEIV 576
Query: 119 DFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNKLVKE 178
DFHGEMVLLENYSALNYTGLVKI+KKYDKRTGAL+RLPFIQ VL QPFF D+L KLVKE
Sbjct: 577 DFHGEMVLLENYSALNYTGLVKILKKYDKRTGALIRLPFIQKVLQQPFFTTDMLYKLVKE 756
Query: 179 CEVMLSIIFPKSGP--LGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQNME 234
CE ML +FP + P +G+ + +E + T+T E+ L +PKE EI+ ME
Sbjct: 757 CETMLDYLFPVNVPPAVGEIIPEAEGCDP---STSTTTESDGLL--IPKELEEIEYME 915
Score = 43.1 bits (100), Expect(2) = 3e-75
Identities = 22/31 (70%), Positives = 24/31 (76%)
Frame = +3
Query: 243 SALDTLKEIRGGSSTVSIYSLPPLHSETLVE 273
SAL LKEIR GSSTVS++SLPPL S L E
Sbjct: 942 SALHVLKEIRSGSSTVSMFSLPPLQSSGLEE 1034
>TC90023 similar to PIR|G84886|G84886 hypothetical protein At2g45130
[imported] - Arabidopsis thaliana, partial (48%)
Length = 757
Score = 187 bits (475), Expect = 4e-48
Identities = 101/201 (50%), Positives = 132/201 (65%), Gaps = 13/201 (6%)
Frame = +3
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEV 60
MKF K L Q++ TLP+WRDKFLSYK+LKK L+LI S + L+ G
Sbjct: 105 MKFGKRLKQQVQDTLPEWRDKFLSYKELKKLLRLI-------SAAPTSLLNGSIG----Y 251
Query: 61 TKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVA-----WAKSSDI------- 108
K +F+ LL EI+KFNGFF+E+EE+++I+ KE+Q ++ W +
Sbjct: 252 GKSEAEFMYLLNNEIDKFNGFFMEQEEDFIIRHKEVQQRIKRVVDLWGPNGSQPSEAEYK 431
Query: 109 -ELMTVGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFF 167
E+ + + IVDFHGEMVLL NYS +NYTGL KI+KKYDKRTG LLRLPFIQ VL QPFF
Sbjct: 432 EEMEKIRKAIVDFHGEMVLLVNYSNINYTGLAKILKKYDKRTGGLLRLPFIQKVLEQPFF 611
Query: 168 KIDVLNKLVKECEVMLSIIFP 188
D+++KLVKECE ++ +FP
Sbjct: 612 TTDLISKLVKECETIIDAVFP 674
>BF650200 similar to GP|15215822|gb AT5g15330/F8M21_220 {Arabidopsis
thaliana}, partial (45%)
Length = 651
Score = 150 bits (378), Expect = 7e-37
Identities = 84/187 (44%), Positives = 119/187 (62%), Gaps = 15/187 (8%)
Frame = +1
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEV 60
MKF K +E+T+P+WRDKFL YK LKK LK +P ++ L +
Sbjct: 97 MKFGKEFKTHLEETIPEWRDKFLCYKPLKKLLKHHLPITTTTTTPINLHLHF---LQQPF 267
Query: 61 TKEVKD--FLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAK---------SSDIE 109
+ + FLR+L E+EKFN F+V+KEEE+VI+++EL++++ K +SD E
Sbjct: 268 SPNILQAWFLRILNQELEKFNDFYVDKEEEFVIRFQELKERIERLKEKSSQSEKYTSDCE 447
Query: 110 ----LMTVGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQP 165
+M + +++V HGEMVLL+NYS+LN+ GL+KI+KKYDKRTG LL+LPF Q VL QP
Sbjct: 448 FSEEMMDIRKDLVTIHGEMVLLKNYSSLNFAGLIKILKKYDKRTGGLLQLPFTQIVLRQP 627
Query: 166 FFKIDVL 172
FF + L
Sbjct: 628 FFTTEPL 648
>CB892908
Length = 231
Score = 133 bits (334), Expect = 8e-32
Identities = 64/77 (83%), Positives = 70/77 (90%)
Frame = +1
Query: 15 LPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEVTKEVKDFLRLLEVE 74
LPDWRD+FLSY+DL QLKLIVP EIDSSCS RRRLDDDGGA+GEVTKEV DFLRLLEV+
Sbjct: 1 LPDWRDRFLSYRDLMMQLKLIVPSEIDSSCSIRRRLDDDGGADGEVTKEVIDFLRLLEVD 180
Query: 75 IEKFNGFFVEKEEEYVI 91
IE FNGFFVE E++YVI
Sbjct: 181IETFNGFFVETEDDYVI 231
>BG451965 similar to GP|16506648|gb| IDS-4-like protein {Castanea sativa},
partial (16%)
Length = 658
Score = 79.7 bits (195), Expect = 1e-15
Identities = 38/38 (100%), Positives = 38/38 (100%)
Frame = +1
Query: 100 VAWAKSSDIELMTVGREIVDFHGEMVLLENYSALNYTG 137
VAWAKSSDIELMTVGREIVDFHGEMVLLENYSALNYTG
Sbjct: 1 VAWAKSSDIELMTVGREIVDFHGEMVLLENYSALNYTG 114
>AL386005 weakly similar to PIR|G84886|G848 hypothetical protein At2g45130
[imported] - Arabidopsis thaliana, partial (13%)
Length = 179
Score = 45.1 bits (105), Expect = 3e-05
Identities = 20/35 (57%), Positives = 27/35 (77%)
Frame = +2
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLI 35
MKF KI + I++ P WRDKFLSYK+LK+ ++LI
Sbjct: 65 MKFAKIYIDDIKEKQPGWRDKFLSYKELKRLIRLI 169
>TC92010
Length = 450
Score = 40.8 bits (94), Expect = 6e-04
Identities = 17/51 (33%), Positives = 33/51 (64%)
Frame = +3
Query: 154 RLPFIQDVLNQPFFKIDVLNKLVKECEVMLSIIFPKSGPLGQSLSTSEVFE 204
RLPF+Q +L PF D++++LV+ECE ++ + ++G + ++ VF+
Sbjct: 6 RLPFLQKILEHPFLSTDLISELVRECENIIDEAY-QAGEAAERVNAKAVFD 155
>TC80935 similar to GP|11994328|dbj|BAB02287. receptor protein-like
{Arabidopsis thaliana}, partial (13%)
Length = 733
Score = 37.7 bits (86), Expect = 0.005
Identities = 34/146 (23%), Positives = 67/146 (45%), Gaps = 46/146 (31%)
Frame = +3
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLI----VPKE-----------IDSSCS 45
+KF K L Q+ +P+W++ F++Y LKKQ+K I +PK+ +S C
Sbjct: 3 VKFSKELEAQL---IPEWKEAFVNYWQLKKQIKRIKLSKIPKQNHNHAEGGGSIFNSLCF 173
Query: 46 KRRRL--------DDDG------------GAEGEVTK-----------EVKDFLRLLEVE 74
+++ D+D G+ E+ + EV+ F +L+ E
Sbjct: 174 HVKKISLKLSPESDNDNTNIIKVRKKTIKGSGEEIYQTELVQLFSEEDEVRVFFAMLDDE 353
Query: 75 IEKFNGFFVEKEEEYVIKWKELQDKV 100
+ K N F++++E E++ + + L ++
Sbjct: 354 LNKVNQFYIKQENEFIERREALNKQL 431
>TC91758 similar to GP|11994328|dbj|BAB02287. receptor protein-like
{Arabidopsis thaliana}, partial (5%)
Length = 353
Score = 34.3 bits (77), Expect = 0.053
Identities = 19/43 (44%), Positives = 28/43 (64%), Gaps = 4/43 (9%)
Frame = +1
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLI----VPKE 39
+KF K L Q+ +P+W+D F++YK LKK +K I VPK+
Sbjct: 106 VKFSKELEAQL---IPEWKDAFVNYKMLKKHIKKIKLSRVPKK 225
>TC79049 weakly similar to PIR|T10798|T10798 pherophorin-S - Volvox carteri,
partial (6%)
Length = 957
Score = 31.2 bits (69), Expect = 0.45
Identities = 32/125 (25%), Positives = 58/125 (45%), Gaps = 27/125 (21%)
Frame = -3
Query: 26 KDLKKQLKLIVPKEIDSSCSKRRRLDDD-GGAEGEVTKEVKDFLRLLEVEIEKFN----- 79
KDL+K++ ++ KEI+ +KR R++++ GE +E+ F +E E+EK
Sbjct: 805 KDLEKRIGVLEMKEIEER-NKRIRVEEELRDTIGEKDREIDGFRNKVE-ELEKVGAEKKD 632
Query: 80 --GFFVEKEEEYVIKWKELQDKVAWAKSSDIELM-------------------TVGREIV 118
G ++ ++ Y +E ++K +S ++L+ VGREI
Sbjct: 631 EAGDWLNEKLSYEKALRESEEKAKGFESQIVQLLEEVGESGKMIKSLNEKAAEIVGREIN 452
Query: 119 DFHGE 123
HGE
Sbjct: 451 GIHGE 437
>TC80700 similar to GP|9759160|dbj|BAB09716.1 contains similarity to myosin
heavy chain~gene_id:MEE6.21 {Arabidopsis thaliana},
partial (4%)
Length = 958
Score = 29.6 bits (65), Expect = 1.3
Identities = 15/42 (35%), Positives = 24/42 (56%)
Frame = +2
Query: 59 EVTKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKV 100
E K ++D ++ E +E ++EKE E K +EL+DKV
Sbjct: 224 EKIKMLEDLIKSKETALEASASSYLEKERELQSKIEELEDKV 349
>TC86630 similar to GP|22136930|gb|AAM91809.1 unknown protein {Arabidopsis
thaliana}, partial (41%)
Length = 1927
Score = 29.3 bits (64), Expect = 1.7
Identities = 28/118 (23%), Positives = 53/118 (44%), Gaps = 8/118 (6%)
Frame = +3
Query: 35 IVPKEIDSSCSKRRRLDDDGGAEGEVTKEVKD-------FLRLLEVEIEKFNGFFVEKEE 87
++PK++ + L G + TK+V+ FLR +E +EK +E+ E
Sbjct: 378 VLPKDLGIGRERNGGLTIVGSYVPKTTKQVEQLKLQCGHFLRSIEASVEKLAMGSIEERE 557
Query: 88 EYVIKWKELQDKVAWAKSSDIELMTVGREIVDFHGEMVLLE-NYSALNYTGLVKIIKK 144
+ + + EL D + K+ L+ R ++ L+ NY + LV+I+K+
Sbjct: 558 DEISRAAELAD--VYLKNRKDTLVMTSRNLITGKSASESLDINYKV--SSALVEIMKR 719
>TC92237 similar to PIR|B96712|B96712 probable receptor protein F14K14.15
[imported] - Arabidopsis thaliana, partial (4%)
Length = 544
Score = 28.9 bits (63), Expect = 2.2
Identities = 10/23 (43%), Positives = 16/23 (69%)
Frame = +2
Query: 13 QTLPDWRDKFLSYKDLKKQLKLI 35
Q +P+W++ F+ Y LKK +K I
Sbjct: 260 QLIPEWKEAFVDYWQLKKDIKRI 328
>BF635781 similar to GP|17104621|gb| unknown protein {Arabidopsis thaliana},
partial (9%)
Length = 585
Score = 28.5 bits (62), Expect = 2.9
Identities = 13/24 (54%), Positives = 16/24 (66%)
Frame = +2
Query: 41 DSSCSKRRRLDDDGGAEGEVTKEV 64
D SKRRRLD+ G GEVT ++
Sbjct: 233 DDVSSKRRRLDNKDGGGGEVTTDL 304
>TC82362 similar to GP|15450519|gb|AAK96552.1 AT4g22540/F7K2_120
{Arabidopsis thaliana}, partial (19%)
Length = 772
Score = 28.1 bits (61), Expect = 3.8
Identities = 15/64 (23%), Positives = 31/64 (48%)
Frame = +1
Query: 192 PLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQNMENIFIKLTTSALDTLKEI 251
P S+ST + + + E ++ N KE + EFS++Q I + + DT++++
Sbjct: 88 PYHISVSTERLKKRLLEEGSSENLVKECEQIMLAEFSDLQEKLQILCQERSGLFDTIRQL 267
Query: 252 RGGS 255
+
Sbjct: 268 EAAN 279
>TC82605 similar to GP|17104721|gb|AAL34249.1 unknown protein {Arabidopsis
thaliana}, partial (56%)
Length = 840
Score = 27.7 bits (60), Expect = 4.9
Identities = 16/54 (29%), Positives = 27/54 (49%), Gaps = 4/54 (7%)
Frame = -2
Query: 51 DDDGGAEGEVTKEVKDFLRLLEVEIEKFNGFFVEKE----EEYVIKWKELQDKV 100
DD+GG+ + R LEVE+E+ +EK+ E V++W + +V
Sbjct: 197 DDEGGSGSGSPWNFSNLARSLEVEVEERREIPLEKDMEEGESVVVEWNWMLQQV 36
>TC78783 similar to GP|21537171|gb|AAM61512.1 unknown {Arabidopsis
thaliana}, partial (21%)
Length = 1431
Score = 27.3 bits (59), Expect = 6.5
Identities = 11/39 (28%), Positives = 22/39 (56%)
Frame = +2
Query: 24 SYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEVTK 62
S++D + K+ DSSC+K +++ + A+ E+ K
Sbjct: 896 SFQDSDRTFSSSESKDSDSSCNKNSKVNSESAAKAELLK 1012
>TC89977 similar to GP|5764395|gb|AAD51282.1| far-red impaired response
protein {Arabidopsis thaliana}, partial (35%)
Length = 1523
Score = 27.3 bits (59), Expect = 6.5
Identities = 9/18 (50%), Positives = 15/18 (83%)
Frame = +2
Query: 84 EKEEEYVIKWKELQDKVA 101
EK+EE+++ WKEL +V+
Sbjct: 329 EKDEEFLVTWKELSSEVS 382
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.136 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,382,742
Number of Sequences: 36976
Number of extensions: 85514
Number of successful extensions: 488
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 480
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 481
length of query: 274
length of database: 9,014,727
effective HSP length: 95
effective length of query: 179
effective length of database: 5,502,007
effective search space: 984859253
effective search space used: 984859253
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC147471.4


