

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147471.11 - phase: 0 /pseudo
(1453 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
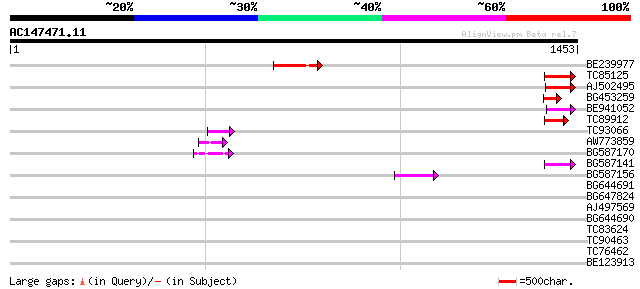
Score E
Sequences producing significant alignments: (bits) Value
BE239977 weakly similar to GP|23237899|db polyprotein-like {Oryz... 139 1e-32
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 69 1e-11
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 65 1e-10
BG453259 homologue to GP|21434|emb|CA ORF4 {Solanum tuberosum}, ... 62 2e-09
BE941052 weakly similar to PIR|B85188|B85 retrotransposon like p... 60 5e-09
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 58 2e-08
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 50 8e-06
AW773859 49 1e-05
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 49 1e-05
BG587141 similar to PIR|H86461|H86 hypothetical protein AAF32440... 47 5e-05
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 45 2e-04
BG644691 similar to GP|21434|emb|CA ORF4 {Solanum tuberosum}, pa... 42 0.002
BG647824 weakly similar to PIR|G96722|G96 hypothetical protein F... 38 0.033
AJ497569 weakly similar to PIR|T04833|T04 hypothetical protein F... 35 0.21
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 34 0.47
TC83624 homologue to PIR|G84581|G84581 copia-like retroelement p... 34 0.47
TC90463 31 4.0
TC76462 similar to GP|10176957|dbj|BAB10277. gene_id:MHJ24.7~unk... 30 5.2
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 30 6.8
>BE239977 weakly similar to GP|23237899|db polyprotein-like {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 514
Score = 139 bits (349), Expect = 1e-32
Identities = 83/128 (64%), Positives = 97/128 (74%), Gaps = 4/128 (3%)
Frame = -1
Query: 677 YGSFILILS*FVYL**SHP*NIWVYVVCPYP**W*RET*S*SFKMCLYRVFFHPKGLQML 736
Y SF ++LS +L *SH *NIWV+VVCPYP**W*+ *S*S K+CLY+VF H KGLQML
Sbjct: 514 YRSFTIVLSLCSHLK*SHS*NIWVHVVCPYP**W*K*I*S*SLKVCLYKVFLHSKGLQML 335
Query: 737 SSS-SQILCLSRCHLP*TRKLLCSNSSSGVVF---M*GR*VSFTS*P*SRTRS*G*NWK* 792
SSS S +LC SRCH P*TRKL S ++F +*GR*+S TS*P R R+*G*NW+*
Sbjct: 334 SSSIS*VLCFSRCHFP*TRKLFWSKL---IIFRG*I*GR*ISSTS*PYFRARN*G*NWR* 164
Query: 793 QC*DGS*F 800
QC*DG *F
Sbjct: 163 QC*DGC*F 140
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 68.9 bits (167), Expect = 1e-11
Identities = 29/79 (36%), Positives = 50/79 (62%)
Frame = +3
Query: 1371 EPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLT 1430
E + +YCD++ A++IA NP H RTKH+ + HF++E ++ G + + + DNLAD +T
Sbjct: 297 EQITVYCDSQSALHIARNPAFHSRTKHIGIQYHFVREVVEEGSVDMQKIHTNDNLADAMT 476
Query: 1431 KRLNNNNFEKFVSKLGMID 1449
K +N + F S G+++
Sbjct: 477 KSINTDKFIWCRSSYGLLE 533
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 65.5 bits (158), Expect = 1e-10
Identities = 29/77 (37%), Positives = 47/77 (60%)
Frame = +2
Query: 1372 PMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTK 1431
P ++YCDNK AI ++ NPV H R+KH+++ H I+E + + Y +++ +AD+ TK
Sbjct: 212 PTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCPTEEKIADIFTK 391
Query: 1432 RLNNNNFEKFVSKLGMI 1448
L +F K LGM+
Sbjct: 392 PLKIESFYKLKKMLGMM 442
>BG453259 homologue to GP|21434|emb|CA ORF4 {Solanum tuberosum}, partial (5%)
Length = 657
Score = 62.0 bits (149), Expect = 2e-09
Identities = 27/47 (57%), Positives = 37/47 (78%)
Frame = -2
Query: 1368 DFDEPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLI 1414
++ +PM L+ +N + IAHNPVQH RTKH+E+++HFI EKL SGLI
Sbjct: 287 NYKDPMTLF*NNNFVSRIAHNPVQHYRTKHIEIDQHFIIEKLYSGLI 147
>BE941052 weakly similar to PIR|B85188|B85 retrotransposon like protein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 480
Score = 60.5 bits (145), Expect = 5e-09
Identities = 27/75 (36%), Positives = 44/75 (58%)
Frame = +2
Query: 1375 LYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTKRLN 1434
L CD A + HNPV H R KH+ ++ HF+++ + G + +V + D LAD LTK L+
Sbjct: 23 LRCDYLSATYLTHNPVYHSRMKHISIDIHFVRDLVQQGKLKVQHVCTVDQLADCLTKPLS 202
Query: 1435 NNNFEKFVSKLGMID 1449
+ + +K+G+ D
Sbjct: 203 KSRHQLLRNKIGVTD 247
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 58.2 bits (139), Expect = 2e-08
Identities = 23/61 (37%), Positives = 46/61 (74%)
Frame = +1
Query: 1371 EPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLT 1430
E ++++CD++ AI++A++ V H+RTKH+++ HFI++ ++S I ++S++N AD+ T
Sbjct: 310 EYVKIHCDSQSAIHLANHQVYHERTKHIDIRLHFIRDMIESKEIVVEKMASEENPADVFT 489
Query: 1431 K 1431
K
Sbjct: 490 K 492
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 49.7 bits (117), Expect = 8e-06
Identities = 24/70 (34%), Positives = 36/70 (51%)
Frame = +1
Query: 507 LDVESFQCELCELAKHKRVTFSVSNKMSTFPFYLVHTNVWGPSNVPNISGARWFVTFIDD 566
+D F L K+V+FS + + +H+++WGPS V + G R+ +T IDD
Sbjct: 4 IDKLEFCKHLLFFGNRKKVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDD 183
Query: 567 CTWVTWVYFL 576
WVYFL
Sbjct: 184 FPRKVWVYFL 213
>AW773859
Length = 538
Score = 49.3 bits (116), Expect = 1e-05
Identities = 25/77 (32%), Positives = 44/77 (56%), Gaps = 1/77 (1%)
Frame = -3
Query: 483 LYHYRVGDPSFRVIKQLFPLF-FKTLDVESFQCELCELAKHKRVTFSVSNKMSTFPFYLV 541
L+H+R+G S R + L F F T+D S C++C ++HK++ F +S ++ + L
Sbjct: 230 LWHFRLGHLSNRKLLSLHSNFPFITIDQNSV-CDICHYSRHKKLPFQLSTNRASKCYELF 54
Query: 542 HTNVWGPSNVPNISGAR 558
H ++WGP + +I R
Sbjct: 53 HFDIWGPFSTQSIHNQR 3
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 48.9 bits (115), Expect = 1e-05
Identities = 32/106 (30%), Positives = 55/106 (51%), Gaps = 2/106 (1%)
Frame = -3
Query: 470 FLESIKTNREKVLLYHYRVGDPSFRVIKQLFPLFFKTLDVESFQCELCELAKHKRVTFSV 529
F S N++ L+H R+G P R + + P + E+ CE C L KH + F
Sbjct: 617 FTSSSSLNKDA--LWHARLGHPHGRALNLMLP----GVVFENKNCEACILGKHCKNVFPR 456
Query: 530 SNKMSTFPFYLVHTNVWGPSNVPNIS--GARWFVTFIDDCTWVTWV 573
++ + F L++T++W P++S ++FVTFID+ + TW+
Sbjct: 455 TSTVYENCFDLIYTDLW---TAPSLSRDNHKYFVTFIDEKSKYTWL 327
>BG587141 similar to PIR|H86461|H86 hypothetical protein AAF32440.1 [imported]
- Arabidopsis thaliana, partial (20%)
Length = 731
Score = 47.0 bits (110), Expect = 5e-05
Identities = 25/80 (31%), Positives = 42/80 (52%)
Frame = +3
Query: 1371 EPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLT 1430
E + + DN+ I + NPV H R H+ HFI+E +++G + +V + + A + T
Sbjct: 186 EEVVIRIDNQSVIALTRNPVFHGRGNHIHKRYHFIRECVENGQVEVEHVPGEKHRAYI*T 365
Query: 1431 KRLNNNNFEKFVSKLGMIDI 1450
K L F + +GMID+
Sbjct: 366 KALGRIIFREIRYYIGMIDL 425
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 45.1 bits (105), Expect = 2e-04
Identities = 30/113 (26%), Positives = 57/113 (49%)
Frame = -3
Query: 987 MGIHCKIQG*WFY*EIQGKIGSQRIHSDLWSRLLRDICPYCKNEYCQGDIIFSS*LQLEL 1046
M ++ ++QG*W *E + S+R++SD+W L DIC + + + +
Sbjct: 424 MDLYNQVQG*WVD*EEEN*TSSKRVYSDIWRGLH*DICTSSQATHN*NCFKLGCEPWMGI 245
Query: 1047 API*CEKCLPSWRT*RRDLHECAPWIR*TYYYQHRVQVEKSFIWAKAVTTCMV 1099
CE+C+ + RT* L+ + + + +V++ ++WA+A+T MV
Sbjct: 244 VANGCEECISTRRT*G*SLYVSSTRS*TSSEERECTEVKEGYLWAEAITKSMV 86
>BG644691 similar to GP|21434|emb|CA ORF4 {Solanum tuberosum}, partial (5%)
Length = 753
Score = 41.6 bits (96), Expect = 0.002
Identities = 25/64 (39%), Positives = 39/64 (60%), Gaps = 4/64 (6%)
Frame = +1
Query: 1378 DNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTP----YVSSQDNLADLLTKRL 1433
DN I+IAHNP+ HDRTKH E+++H + + + TP +V S +N +LTK L
Sbjct: 445 DNIIPISIAHNPI*HDRTKHTEIDRHLHQRE----SLVTP*VLLFVQSINN-QRMLTKGL 609
Query: 1434 NNNN 1437
+ ++
Sbjct: 610 SKSS 621
>BG647824 weakly similar to PIR|G96722|G96 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (5%)
Length = 721
Score = 37.7 bits (86), Expect = 0.033
Identities = 19/42 (45%), Positives = 28/42 (66%), Gaps = 1/42 (2%)
Frame = -3
Query: 1369 FDEPMRLYCDNKYAI-NIAHNPVQHDRTKHLEVNKHFIKEKL 1409
F +P LYCDN+ A +IA N +RTKH+E++ H ++ KL
Sbjct: 302 FIKPAMLYCDNQSAARHIAANSSFLERTKHIELDCHIVRVKL 177
>AJ497569 weakly similar to PIR|T04833|T04 hypothetical protein F21P8.50 -
Arabidopsis thaliana, partial (4%)
Length = 723
Score = 35.0 bits (79), Expect = 0.21
Identities = 22/59 (37%), Positives = 34/59 (57%)
Frame = +2
Query: 1376 YCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTKRLN 1434
YCDN A++IA N V H+RT H E + + ++ S ++ +S+D A LTK L+
Sbjct: 290 YCDNISALHIAANMVFHERT*HRETDPYIVQ---GSRMLQLMPSASKDQPAYSLTKPLH 457
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 33.9 bits (76), Expect = 0.47
Identities = 35/136 (25%), Positives = 57/136 (41%)
Frame = -1
Query: 1005 KIGSQRIHSDLWSRLLRDICPYCKNEYCQGDIIFSS*LQLELAPI*CEKCLPSWRT*RRD 1064
++G RI S +RL C+N F ++ P CE+C+ WR+ R
Sbjct: 407 QVGGARIQSKRRNRL**GFFTCCQNGSY*NFNSFCCIHGVQAVPNGCEECIY*WRSQRGG 228
Query: 1065 LHECAPWIR*TYYYQHRVQVEKSFIWAKAVTTCMVW*IH*SYGRFGLQRKSRRPYFICQT 1124
+ + WI + VQ+E IW++A + MV Q++ R Y +
Sbjct: 227 VCQATSWI*RCRGTKSCVQIE*DTIWSEASSKSMV*KAVKVSAEEWFQKRQDRQYPVLIK 48
Query: 1125 LRNRGSNSVAGICG*H 1140
R R + + +CG*H
Sbjct: 47 KRIRIAYH-SSVCG*H 3
>TC83624 homologue to PIR|G84581|G84581 copia-like retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(1%)
Length = 831
Score = 33.9 bits (76), Expect = 0.47
Identities = 15/31 (48%), Positives = 20/31 (64%)
Frame = +2
Query: 1375 LYCDNKYAINIAHNPVQHDRTKHLEVNKHFI 1405
+YC N+ + IA N V H+RTKH E N F+
Sbjct: 482 IYCVNQITLYIAKNQVYHERTKH*ENNWTFL 574
>TC90463
Length = 1175
Score = 30.8 bits (68), Expect = 4.0
Identities = 14/27 (51%), Positives = 18/27 (65%)
Frame = -2
Query: 1425 LADLLTKRLNNNNFEKFVSKLGMIDIH 1451
L D LTK L F F+SKLGM++I+
Sbjct: 322 LPDFLTKALPPPKFHSFISKLGMLNIY 242
>TC76462 similar to GP|10176957|dbj|BAB10277. gene_id:MHJ24.7~unknown
protein {Arabidopsis thaliana}, partial (34%)
Length = 1154
Score = 30.4 bits (67), Expect = 5.2
Identities = 19/47 (40%), Positives = 26/47 (54%)
Frame = +2
Query: 339 PFLVNFHFLLNLMLQIHPLNNTGY*ILEPLTT*HLYLHTFLPIHLVL 385
P LV FH LLNL L ++PLN P ++ L++ T L H+ L
Sbjct: 197 PLLV*FHLLLNLTLLLNPLN-------VPPSSVSLFILTSLRSHICL 316
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 30.0 bits (66), Expect = 6.8
Identities = 31/117 (26%), Positives = 52/117 (43%), Gaps = 5/117 (4%)
Frame = +1
Query: 1148 R*AKTVRPTLSQRVRDQDLGKIEVFFGN*SGSF*ERNFHISTEIHYRSSVGDW--QDSMQ 1205
+* K ++ L++ +DLG ++ F G + + I R V D + M
Sbjct: 166 K*IKRLKNLLAEEFEIKDLGNLKYFLG-----MEVARWKKGSSISQRKYVLDLLKETRMI 330
Query: 1206 ACK*TN*S---KCEVGECRRRCCS**RNVSKVSWQAYIPLTY*T*CCF*CKLS*PIH 1259
CK CE R+ S**R +SKV W+ ++ +++*T F + P+H
Sbjct: 331 GCKTIRDPYGCNCEARNSRQWDTS**RKISKVGWKTHLFISH*TRY*FCSVYNEPVH 501
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.364 0.163 0.626
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 51,349,222
Number of Sequences: 36976
Number of extensions: 826818
Number of successful extensions: 11117
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 2497
Number of HSP's successfully gapped in prelim test: 405
Number of HSP's that attempted gapping in prelim test: 8382
Number of HSP's gapped (non-prelim): 3462
length of query: 1453
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1345
effective length of database: 5,021,319
effective search space: 6753674055
effective search space used: 6753674055
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 36 (21.5 bits)
S2: 65 (29.6 bits)
Medicago: description of AC147471.11


