

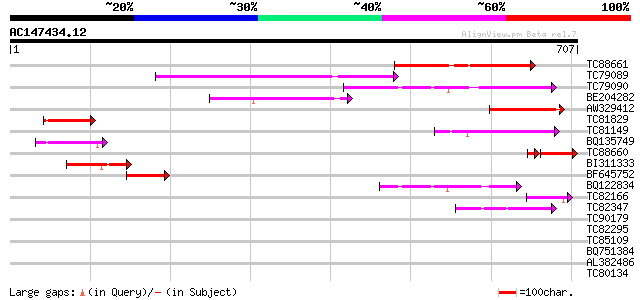
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147434.12 - phase: 0
(707 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88661 weakly similar to GP|22507085|gb|AAM97760.1 putative tra... 174 1e-43
TC79089 weakly similar to PIR|T46111|T46111 probable transposase... 151 7e-37
TC79090 similar to PIR|T46111|T46111 probable transposase - Arab... 135 7e-32
BE204282 weakly similar to PIR|C85069|C850 hypothetical protein ... 98 1e-20
AW329412 weakly similar to GP|11994591|db Ac transposase-like pr... 94 1e-19
TC81829 91 2e-18
TC81149 weakly similar to GP|11994591|dbj|BAB02646. Ac transposa... 90 3e-18
BQ135749 similar to GP|16944417|emb hypothetical protein {Neuros... 82 7e-16
TC88660 PIR|T48044|T48044 hypothetical protein T12C14.220 - Arab... 68 4e-15
BI311333 79 6e-15
BF645752 homologue to GP|22713420|gb| Unknown (protein for MGC:3... 64 2e-10
BQ122834 weakly similar to GP|20521440|dbj hypothetical protein~... 48 1e-05
TC82166 similar to GP|10140707|gb|AAG13541.1 putative Tam3-trans... 47 2e-05
TC82347 47 3e-05
TC90179 weakly similar to GP|9719456|gb|AAF97810.1| transposase ... 42 0.001
TC82295 similar to PIR|A96722|A96722 unknown protein T17F3.2 [im... 41 0.002
TC85109 33 0.49
BQ751384 32 1.1
AL382486 32 1.1
TC80134 GP|7295572|gb|AAF50883.1| shakB gene product {Drosophila... 31 1.9
>TC88661 weakly similar to GP|22507085|gb|AAM97760.1 putative transposase
{Oryza sativa (japonica cultivar-group)}, partial (4%)
Length = 521
Score = 174 bits (441), Expect = 1e-43
Identities = 89/176 (50%), Positives = 118/176 (66%), Gaps = 1/176 (0%)
Frame = +1
Query: 481 YNKYWGKIENINKLIYFGVILDPRYKFSYVEWCFNDMYGDQPTFFTDLIAMIHTQLFKLF 540
Y+KYWG + +N+ +YFGVI DPRYKF Y+EW FND+YG + + LFKL+
Sbjct: 1 YDKYWGNVVKMNQFLYFGVIFDPRYKFGYIEWSFNDLYGAGSDIAKERAGSVSDNLFKLY 180
Query: 541 NWYKDAYDQQHNSGHPSASPSESSYVSENVIPAEVPSHLARAEAFKEHLKLKESIVKKNE 600
N YK ++ S S +S V + +P ++PS A+A+K+HLK KE+IV +N+
Sbjct: 181 NLYKSEHE-----SFVGPSGSNNSSVEQPAVP-KIPSLNTTADAYKKHLKTKETIVPQND 342
Query: 601 LERYL-DEERAEDVNFDILLSWKQNSCRYPVLSSMVRDVLATPVSTVASESAFSTG 655
LERYL D +D +FDIL WK+N RYPVL++MVRDVLATPVS+VASESA STG
Sbjct: 343 LERYLSDPPENDDPSFDILTWWKKNCVRYPVLATMVRDVLATPVSSVASESAXSTG 510
>TC79089 weakly similar to PIR|T46111|T46111 probable transposase -
Arabidopsis thaliana, partial (36%)
Length = 942
Score = 151 bits (382), Expect = 7e-37
Identities = 90/303 (29%), Positives = 143/303 (46%), Gaps = 1/303 (0%)
Frame = +3
Query: 183 TVARDCFQLFLDENLRLKTYFKSDCVRVALTTDCWTSGQNFSYMTLTAHFINNDWKYEKR 242
T+ DC +L E L YF R LT D WTS Q+ Y+ +T HF+++DWK +KR
Sbjct: 33 TIQGDCVATYLTEKQNLSKYFDELPGRFGLTLDMWTSSQSVGYVFITGHFVDSDWKLQKR 212
Query: 243 ILSFCTVPNHKGDT-IGRKVEEILKEWGIRNVSTITVDNASSNDVAVAYLKKRINNMGGL 301
IL+ P D+ + V + +W + N VA+ L+ ++ L
Sbjct: 213 ILNVVMEPCPDSDSALSHAVSACISDWNLEGRLFTITCNQPLTKVALENLRPLLSVKNPL 392
Query: 302 MGDGSFFHLRCCAHILNLVVGDGLKQNELSISSIRNSVRFVRSSPQRSAKFKECIEFARI 361
+ +G C A L+ V D L + ++ IR SV++V++S KF + E ++
Sbjct: 393 IFNGQLLIGHCIARTLSNVAYDLLSSAQGIVNKIRESVKYVKTSESHEDKFLDLKEHLQV 572
Query: 362 TCKKMLCLDVQTRWNTAYLMLDGAEKFQPAFEKLEGEDSGYLEFFGEAGPPSIHDWENVR 421
+K L +D QT+WNT Y ML A + + F L+ D Y G PSI DW+ V
Sbjct: 573 PSEKSLFIDDQTKWNTTYQMLVAASELKEVFSCLDTSDPDY------KGAPSIPDWKLVE 734
Query: 422 CFVRFLKIFYDATKEFSSSQGVSLHKAFHQLASVHCELKRSAMNLNTVLASMGSDMKQKY 481
+LK YDA ++ + FH++ +H +L R+ N + ++ + M K
Sbjct: 735 ILCTYLKPLYDAANILMTTTYPTAITFFHEVXKLHLDLSRAXKNEDPFISDLTKPMYXKI 914
Query: 482 NKY 484
+KY
Sbjct: 915 DKY 923
>TC79090 similar to PIR|T46111|T46111 probable transposase - Arabidopsis
thaliana, partial (28%)
Length = 1068
Score = 135 bits (339), Expect = 7e-32
Identities = 81/276 (29%), Positives = 142/276 (51%), Gaps = 10/276 (3%)
Frame = +2
Query: 417 WENVRCFVRFLKIFYDATKEFSSSQGVSLHKAFHQLASVHCELKRSAMNLNTVLASMGSD 476
W+ V +LK YDA +++ + FH++ +H +L R+ N + ++ +
Sbjct: 56 WKLVEILCTYLKPLYDAANILTTTTYPTAITFFHEVWKLHLDLARAVKNEDPFISDLTKP 235
Query: 477 MKQKYNKYWGKIENINKLIYFGVILDPRYKFSYVEWCFNDMYGDQPTFFTDLIAMIHTQL 536
M +K +KYW + + + V++DPR+K VE+ F +YG+ + + ++ +
Sbjct: 236 MYEKIDKYW---RDCSLALVIAVVMDPRFKMKLVEFSFTKIYGEDAHAY---VKIVDDGI 397
Query: 537 FKLFNWYKD-------AYDQQHNSGHPSASPSES--SYVSENVIPAEVPSHLARAEAFKE 587
+LF+ Y AY ++ N+G S + +S+N + F
Sbjct: 398 HELFHEYATLPLPLTPAYAEEGNAGSSMKMEGSSGGTLLSDNGLTD-----------FDA 544
Query: 588 HLKLKESIVKKNELERYLDEERAEDV-NFDILLSWKQNSCRYPVLSSMVRDVLATPVSTV 646
++ + K+EL++YL+E V +FD+L WK N +YP LS M RD+L+ PVSTV
Sbjct: 545 YIMETSTHQTKSELDQYLEESLLPRVPDFDVLGWWKLNKLKYPTLSKMARDILSIPVSTV 724
Query: 647 ASESAFSTGGRVLDTYRSSLNPQMAEALICAQNWLK 682
S+S F + +D YRSSL P+ EAL+CA++W++
Sbjct: 725 PSDSIFDKKSKEMDQYRSSLRPETVEALVCAKDWMQ 832
>BE204282 weakly similar to PIR|C85069|C850 hypothetical protein AT4g05510
[imported] - Arabidopsis thaliana, partial (16%)
Length = 563
Score = 97.8 bits (242), Expect = 1e-20
Identities = 65/188 (34%), Positives = 97/188 (51%), Gaps = 10/188 (5%)
Frame = +3
Query: 250 PNHKGDTIGRKVEEILKEWGI-RNVSTITVDNASSNDVAVAYLKKRINNMGGLM------ 302
P+H + RKV L+EWGI +N+ +IT+DNAS+ND LK ++ ++ L+
Sbjct: 12 PSHDNFELSRKVIGYLQEWGIEKNIFSITMDNASANDGNSQNLKDQLCSLDSLLRTFITS 191
Query: 303 --GDGSFFHLRCCAHILNLVVGDGLKQNELSISSIRNSVRFVRSSPQRSAKFKECI-EFA 359
H +CCAH+L+L+V + LK + IRNS++FV S R +F +C+ E
Sbjct: 192 ATAPSLEKHFKCCAHVLDLMVQESLKVVSDVLDKIRNSLKFVSVSNSRLKQFCQCVEEVG 371
Query: 360 RITCKKMLCLDVQTRWNTAYLMLDGAEKFQPAFEKLEGEDSGYLEFFGEAGPPSIHDWEN 419
L LDV +W + YLML A K++ AFE + +D+ Y PS +WE
Sbjct: 372 GGDSSDSLHLDVSGKWVSTYLMLKSAIKYRSAFEHMCLKDTTYNHC------PSSEEWER 533
Query: 420 VRCFVRFL 427
FL
Sbjct: 534 GEKICEFL 557
>AW329412 weakly similar to GP|11994591|db Ac transposase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 569
Score = 94.4 bits (233), Expect = 1e-19
Identities = 49/95 (51%), Positives = 69/95 (72%), Gaps = 1/95 (1%)
Frame = +3
Query: 599 NELERYLDEER-AEDVNFDILLSWKQNSCRYPVLSSMVRDVLATPVSTVASESAFSTGGR 657
++L++YL+E + +F+IL WK ++ RYP+LS M RDVL TP+ST+A E AF+TGGR
Sbjct: 117 SDLDKYLEEPIFPRNSDFNILNWWKVHTPRYPILSMMARDVLGTPMSTLAPELAFNTGGR 296
Query: 658 VLDTYRSSLNPQMAEALICAQNWLKPTLNQFKDLN 692
+LD+ RSSLNP EALIC +WL+ N+ + LN
Sbjct: 297 LLDSSRSSLNPDTREALICTHDWLR---NESEGLN 392
>TC81829
Length = 701
Score = 90.9 bits (224), Expect = 2e-18
Identities = 40/65 (61%), Positives = 47/65 (71%)
Frame = +2
Query: 43 PLPCLAKRRKPNAGGPRRTSPAWDHFIKLPDEPEPTAACIHCHKRYLCDPKTHGTSNLLA 102
PLP + R+PNA G RRTSP W F LP+EPEP A C HCHKRY C K HGTS++LA
Sbjct: 509 PLP--TRGRRPNANGNRRTSPVWLDFNPLPNEPEPIAVCKHCHKRYRCHSKAHGTSSMLA 682
Query: 103 HSKVC 107
H+K+C
Sbjct: 683 HTKIC 697
>TC81149 weakly similar to GP|11994591|dbj|BAB02646. Ac transposase-like
protein {Arabidopsis thaliana}, partial (11%)
Length = 806
Score = 90.1 bits (222), Expect = 3e-18
Identities = 60/164 (36%), Positives = 94/164 (56%), Gaps = 8/164 (4%)
Frame = +1
Query: 530 AMIHTQLFKLFNWYKDAYDQQHNSGHPSASPSES-SYVSEN-----VIPAEVPSHLARAE 583
+M +++ K+F K Y++ H+ G P AS + ++ EN +P R
Sbjct: 37 SMSASRIEKVFEGVKALYNE-HSIGSPLASHDQGLAWQVENGSSSLSLPWSAKDSRDRLM 213
Query: 584 AFKEHL-KLKESIVKKNELERYLDEER-AEDVNFDILLSWKQNSCRYPVLSSMVRDVLAT 641
F + L + + K++L++YL+E +V+F+IL WK ++ RYPVLS M R+VL
Sbjct: 214 GFDKFLHETSQGEGAKSDLDKYLEEPLFPRNVDFNILNWWKVHTPRYPVLSMMARNVLGI 393
Query: 642 PVSTVASESAFSTGGRVLDTYRSSLNPQMAEALICAQNWLKPTL 685
P+S VA E AF+ GRVLD SSLNP +AL+C+Q+W++ L
Sbjct: 394 PMSKVAPELAFNYSGRVLDRDWSSLNPATVQALVCSQDWIRSEL 525
>BQ135749 similar to GP|16944417|emb hypothetical protein {Neurospora
crassa}, partial (8%)
Length = 533
Score = 82.0 bits (201), Expect = 7e-16
Identities = 42/92 (45%), Positives = 53/92 (56%), Gaps = 3/92 (3%)
Frame = +2
Query: 33 ATQVPVVGLPPLPCLAKRRKPNAGGPRRTSPAWDHFIKLPDEPEPTAACIHCHKRYLCDP 92
+TQ LPP P + ++PNA G RRTS W F LP+E PT AC HCHKRY CD
Sbjct: 260 STQQAAAELPPFPT-TRCKRPNANGNRRTSSVWLDFNPLPNERXPTTACKHCHKRYRCDS 436
Query: 93 KTHGTSNLLAHSKVCF---KNPQNDPTQASLM 121
K +G S++L H +N NDP Q +L+
Sbjct: 437 KLNGKSSMLTHXXXYLXKSQNMLNDPRQTNLI 532
>TC88660 PIR|T48044|T48044 hypothetical protein T12C14.220 - Arabidopsis
thaliana, partial (2%)
Length = 865
Score = 68.2 bits (165), Expect(2) = 4e-15
Identities = 31/46 (67%), Positives = 38/46 (82%)
Frame = +3
Query: 662 YRSSLNPQMAEALICAQNWLKPTLNQFKDLNINEEFELSTTVVSGI 707
YRSSL+P+MAEALIC QNWLKP+ FKDLN++EE+EL VV+ I
Sbjct: 54 YRSSLSPEMAEALICTQNWLKPSFVDFKDLNLSEEYELLENVVAEI 191
Score = 31.6 bits (70), Expect(2) = 4e-15
Identities = 14/15 (93%), Positives = 15/15 (99%)
Frame = +1
Query: 646 VASESAFSTGGRVLD 660
VASESAFSTGGR+LD
Sbjct: 7 VASESAFSTGGRILD 51
>BI311333
Length = 706
Score = 79.0 bits (193), Expect = 6e-15
Identities = 39/83 (46%), Positives = 53/83 (62%), Gaps = 3/83 (3%)
Frame = +2
Query: 72 PDEPEPTAACIHCHKRYLCDPKTHGTSNLLAHSKVCFKNPQ---NDPTQASLMFSNGEGG 128
P +PT AC HCHKRY CD K +G S++LAH +V K P+ NDP Q +L+ NG+G
Sbjct: 14 PMNEKPTTACKHCHKRYRCDSKLNGKSSMLAHCRVYLKKPENMLNDPRQTNLI--NGKGD 187
Query: 129 TLVAASQRFNPAACRKAIALFVL 151
LV+ SQR+N CR I + ++
Sbjct: 188 FLVSVSQRYNAKDCRDFIYVILI 256
>BF645752 homologue to GP|22713420|gb| Unknown (protein for MGC:33586) {Homo
sapiens}, partial (1%)
Length = 321
Score = 64.3 bits (155), Expect = 2e-10
Identities = 31/54 (57%), Positives = 38/54 (69%)
Frame = +3
Query: 146 IALFVLLDEHAFRVVEGEGFKLLCRQLQPLLTIPSRRTVARDCFQLFLDENLRL 199
IA V+L + F VVE + F LC+ LQPL IPSRRTVARDCF++F DE +L
Sbjct: 153 IARLVILSKLPFSVVESDRFNRLCKLLQPLWNIPSRRTVARDCFRMFXDEKFKL 314
>BQ122834 weakly similar to GP|20521440|dbj hypothetical protein~similar to
transposase {Oryza sativa (japonica cultivar-group)},
partial (10%)
Length = 553
Score = 47.8 bits (112), Expect = 1e-05
Identities = 41/189 (21%), Positives = 84/189 (43%), Gaps = 12/189 (6%)
Frame = +2
Query: 462 SAMNLNTVLASMGSDMKQKYNKYWGKIENINKLIYFGVILDPRYKFSYVEWCFNDMYGDQ 521
+A + + L+S+ + + +++YW + ++ V +DPR+K VE F ++G+
Sbjct: 8 AAFSQDPFLSSLIFPLLKNFDQYW---RDSCLILAVAVAMDPRHKMKLVESTFTTIFGEN 178
Query: 522 PTFFTDLIAMIHTQLFKLFNWYK-----------DAYDQQHNSGHPSASPSESSYVSENV 570
+ I ++ L +LF Y D D+ + P P + S +
Sbjct: 179 AEPW---IRIVEDGLHELFIDYNTEMLHFTATNGDDVDEIMLNTEPYEGPVDGSLFVDEG 349
Query: 571 IPAEVPSHLARAEAFKEHLKLKESIVKKNELERYLDEER-AEDVNFDILLSWKQNSCRYP 629
++ +++ + ++ K+E++ YL+E +E FDIL W++N +YP
Sbjct: 350 GLSDFEFNISDFTSMQQF---------KSEMDEYLEEPLLSESKEFDILSWWRENRSKYP 502
Query: 630 VLSSMVRDV 638
LS D+
Sbjct: 503 TLSRAASDI 529
>TC82166 similar to GP|10140707|gb|AAG13541.1 putative Tam3-transposase
{Oryza sativa}, partial (6%)
Length = 574
Score = 47.4 bits (111), Expect = 2e-05
Identities = 26/67 (38%), Positives = 38/67 (55%), Gaps = 9/67 (13%)
Frame = +1
Query: 645 TVASESAFSTGGRVLDTYRSSLNPQMAEALICAQNWLKPTLNQFK---------DLNINE 695
TVASESAFST GR + RS L EAL+C+QNW + + + D ++++
Sbjct: 7 TVASESAFSTSGRDVTPQRSRLKEDTLEALMCSQNWFRTEMQGYSKIYASFECVDEDMDD 186
Query: 696 EFELSTT 702
+ E+S T
Sbjct: 187 DEEMSQT 207
>TC82347
Length = 750
Score = 46.6 bits (109), Expect = 3e-05
Identities = 32/127 (25%), Positives = 59/127 (46%)
Frame = +1
Query: 556 PSASPSESSYVSENVIPAEVPSHLARAEAFKEHLKLKESIVKKNELERYLDEERAEDVNF 615
P+A+ ++ ++ S + P+ S + + H + + + +EL++Y + A+ V+
Sbjct: 208 PAATSAQGTFTSISTTPSVEDSG*TQWLNNRAH---QVAANEASELDQYYRQTIAQPVDN 378
Query: 616 DILLSWKQNSCRYPVLSSMVRDVLATPVSTVASESAFSTGGRVLDTYRSSLNPQMAEALI 675
+ W + +P LS + D+ A P E AFS L + R S+ Q E
Sbjct: 379 PVQW-WISHRKTFPTLSRLALDIFAIPAMATDCERAFSLAKLTLTSQRLSMKSQTLEEAQ 555
Query: 676 CAQNWLK 682
C QNWL+
Sbjct: 556 CLQNWLR 576
>TC90179 weakly similar to GP|9719456|gb|AAF97810.1| transposase
{Cryphonectria parasitica}, partial (3%)
Length = 810
Score = 41.6 bits (96), Expect = 0.001
Identities = 21/92 (22%), Positives = 46/92 (49%), Gaps = 1/92 (1%)
Frame = -1
Query: 180 SRRTVARDCFQLFLDENLRLKTYFKSDCVRVALTTDCWTSGQNFSYMTLTAHFINNDWKY 239
S ++ AR+ QL+ + + +K + +V L+ D WTS + + + HFI+
Sbjct: 408 SDKSQARELHQLYEAKRVIVKEEMRQALTKVHLSFDLWTSPNRLAIIAVFGHFISPAGND 229
Query: 240 EKRILSFCTVPN-HKGDTIGRKVEEILKEWGI 270
+ +L+ P H G+ I + +++++W +
Sbjct: 228 SRYLLALRRQPGAHSGENIASTICQVIRDWDL 133
>TC82295 similar to PIR|A96722|A96722 unknown protein T17F3.2 [imported] -
Arabidopsis thaliana, partial (17%)
Length = 698
Score = 40.8 bits (94), Expect = 0.002
Identities = 36/168 (21%), Positives = 63/168 (37%), Gaps = 34/168 (20%)
Frame = +1
Query: 59 RRTSPAWDHFIKLPDEPEPTAACIHCHKRYLCDPKTHGTSNLLAHSKVCFK--------- 109
R S W+ F ++ A C HC K+ L T GTS+L H C +
Sbjct: 151 RLKSVVWNDFDRIKKGDTCVAVCRHCKKK-LSGSSTSGTSHLRNHLIRCQRRSNHGLAQY 327
Query: 110 ---------------------NPQNDPTQA--SLMFSNGE--GGTLVAASQRFNPAACRK 144
+P D T + ++ F + ++ + F+ R
Sbjct: 328 ITAREKRKEGSLAITNFSLDQDPNKDDTVSLVNIKFEQAQLKDESVNTGNSNFDQRRSRF 507
Query: 145 AIALFVLLDEHAFRVVEGEGFKLLCRQLQPLLTIPSRRTVARDCFQLF 192
+A ++L + +VE GF+ + LQPL + + V DC +++
Sbjct: 508 DLARMIILHGYPLAMVEHVGFRAFVKNLQPLFELVTLNRVEADCIEIY 651
>TC85109
Length = 559
Score = 32.7 bits (73), Expect = 0.49
Identities = 13/43 (30%), Positives = 25/43 (57%)
Frame = +3
Query: 65 WDHFIKLPDEPEPTAACIHCHKRYLCDPKTHGTSNLLAHSKVC 107
W++F+K+ + + C+ C + + CD ++ G S+L H VC
Sbjct: 153 WNYFVKVHKDGKEMCECMTCGRVFTCDGRS-GNSHLNLHIPVC 278
>BQ751384
Length = 455
Score = 31.6 bits (70), Expect = 1.1
Identities = 22/87 (25%), Positives = 42/87 (47%), Gaps = 4/87 (4%)
Frame = -1
Query: 156 AFRVVEGEGFKLLCR---QLQPLLTIPSRRTVARDCFQLFLDE-NLRLKTYFKSDCVRVA 211
+F +++ + FK L + P+L + + + FL N L+ KS + +
Sbjct: 263 SFSILDSKSFKDLLNYYNKSNPILNRHKVKNLLEFTYSRFLSTINYELEANIKS-LGKFS 87
Query: 212 LTTDCWTSGQNFSYMTLTAHFINNDWK 238
LT D WTS +Y+++ +IN+ +K
Sbjct: 86 LTLDIWTSKSQDTYLSIIISYINSSFK 6
>AL382486
Length = 492
Score = 31.6 bits (70), Expect = 1.1
Identities = 22/70 (31%), Positives = 36/70 (51%)
Frame = +3
Query: 605 LDEERAEDVNFDILLSWKQNSCRYPVLSSMVRDVLATPVSTVASESAFSTGGRVLDTYRS 664
LD+ R E NF W + P+LS++ D + P+S+ E +FS +LD R
Sbjct: 231 LDDTRIE--NF-----WLGMVNQLPLLSNIALDYIWLPISSCTVERSFSIYNTILDDDRQ 389
Query: 665 SLNPQMAEAL 674
+L+ + +AL
Sbjct: 390 NLSLESLKAL 419
>TC80134 GP|7295572|gb|AAF50883.1| shakB gene product {Drosophila
melanogaster}, partial (2%)
Length = 828
Score = 30.8 bits (68), Expect = 1.9
Identities = 26/90 (28%), Positives = 44/90 (48%), Gaps = 2/90 (2%)
Frame = -3
Query: 546 AYDQQHNSGHPSASPSESSYVSENV--IPAEVPSHLARAEAFKEHLKLKESIVKKNELER 603
++ ++ H S SPS SSY SE + I A V L R + + L + N+L+
Sbjct: 709 SFSASSHNNHVSPSPSFSSYSSETLAEIAARVIDEL-RWDPHSDEDALYQPWEDDNKLKT 533
Query: 604 YLDEERAEDVNFDILLSWKQNSCRYPVLSS 633
D E+ ED F+ + ++S + V+S+
Sbjct: 532 QNDNEKDEDSEFEFAVV-SRDSTNFSVVSA 446
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.134 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,646,973
Number of Sequences: 36976
Number of extensions: 355445
Number of successful extensions: 1908
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 1881
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1898
length of query: 707
length of database: 9,014,727
effective HSP length: 103
effective length of query: 604
effective length of database: 5,206,199
effective search space: 3144544196
effective search space used: 3144544196
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147434.12


