

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147431.3 + phase: 0 /pseudo
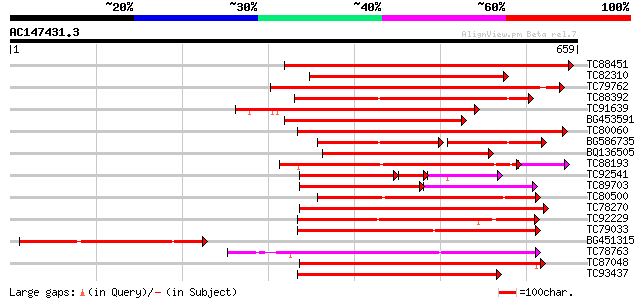
(659 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88451 similar to GP|836954|gb|AAC23542.1|| receptor protein ki... 357 8e-99
TC82310 similar to PIR|T05181|T05181 S-receptor kinase (EC 2.7.1... 338 4e-93
TC79762 similar to PIR|H96731|H96731 hypothetical protein F5A18.... 320 8e-88
TC88392 similar to GP|11275527|dbj|BAB18292. putative receptor-l... 296 1e-80
TC91639 similar to PIR|T05181|T05181 S-receptor kinase (EC 2.7.1... 290 9e-79
BG453591 similar to PIR|T05181|T05 S-receptor kinase (EC 2.7.1.-... 280 1e-75
TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.... 275 3e-74
BG586735 similar to GP|11275527|db putative receptor-like protei... 175 7e-72
BQ136505 similar to PIR|T05754|T05 S-receptor kinase (EC 2.7.1.-... 264 9e-71
TC88193 similar to PIR|G96602|G96602 probable receptor protein k... 254 2e-69
TC92541 similar to PIR|T02153|T02153 protein kinase homolog T1F1... 153 5e-67
TC89703 similar to GP|11994595|dbj|BAB02650. receptor-like serin... 170 3e-65
TC80500 similar to PIR|G96602|G96602 probable receptor protein k... 240 1e-63
TC78270 similar to GP|16649103|gb|AAL24403.1 Unknown protein {Ar... 236 3e-62
TC92229 similar to PIR|B86369|B86369 hypothetical protein AAC980... 233 1e-61
TC79033 similar to GP|21554229|gb|AAM63304.1 somatic embryogenes... 228 6e-60
BG451315 weakly similar to PIR|D85065|D850 receptor protein kina... 226 2e-59
TC78763 somatic embryogenesis receptor kinase 1 [Medicago trunca... 224 6e-59
TC87048 similar to GP|8778594|gb|AAF79602.1| F5M15.3 {Arabidopsi... 223 2e-58
TC93437 similar to GP|11227578|emb|CAC16506. unnamed protein pro... 219 2e-57
>TC88451 similar to GP|836954|gb|AAC23542.1|| receptor protein kinase
{Ipomoea trifida}, partial (28%)
Length = 1276
Score = 357 bits (916), Expect = 8e-99
Identities = 177/337 (52%), Positives = 241/337 (70%), Gaps = 1/337 (0%)
Frame = +3
Query: 320 QDSFNG-ELPTIPLTIIEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQ 378
+D F G ++ T I ++T +FS KLG+GG+GPVYKG L G+E+AVKRLS+TS Q
Sbjct: 159 EDDFKGHDIKVFNFTSILEATMEFSPENKLGQGGYGPVYKGILATGQEIAVKRLSKTSGQ 338
Query: 379 GSEEFKNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKHLDW 438
G EFKNE++ I +LQH+NL +LLG CI +E+IL+YEYMPN SLDF+LF+ K K LDW
Sbjct: 339 GIVEFKNELLLICELQHKNLVQLLGCCIHEEERILIYEYMPNKSLDFYLFDCTKKKLLDW 518
Query: 439 KLRLSIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQ 498
K R +II GIA+GLLYLH+ SRL++IHRDLKASN+LLD+ MNPKI+DFG+AR F + +
Sbjct: 519 KKRFNIIEGIAQGLLYLHKYSRLKIIHRDLKASNILLDENMNPKIADFGMARMFTQQESV 698
Query: 499 TKTKRVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLY 558
T R+ GTYGYM+PEYAM G+ S KSDV+SFGVL+LEI+ G +N F+ + +L+ +
Sbjct: 699 VNTNRIVGTYGYMSPEYAMEGVCSTKSDVYSFGVLLLEIVCGIKNNSFYDVDRPLNLIGH 878
Query: 559 TWKLWCEGKCLELIDPFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRMLGSDTV 618
W+LW +G+ L+L+DP T++ EV +CIH+GLLCV++ A DRPTMS V+ +L + V
Sbjct: 879 AWELWNDGEYLKLMDPTLNDTFVPDEVKRCIHVGLLCVEQYANDRPTMSEVISVLTNKYV 1058
Query: 619 DLPKPTQPAFSVGRKSKNEDQISKNSKDNSVDEETIT 655
P +PAF V R+ + SK ++ TI+
Sbjct: 1059LTNLPRKPAFYVRREIFEGETTSKGQDTDTYSTTTIS 1169
>TC82310 similar to PIR|T05181|T05181 S-receptor kinase (EC 2.7.1.-)
T6K22.120 precursor - Arabidopsis thaliana, partial
(24%)
Length = 696
Score = 338 bits (867), Expect = 4e-93
Identities = 166/231 (71%), Positives = 192/231 (82%)
Frame = +1
Query: 349 GEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQHRNLAKLLGYCIEG 408
G+GGFG VYKG L D +E+AVKRLS+TSSQG EE KNE+I IAKLQHRNL +LL CIE
Sbjct: 1 GKGGFGTVYKGVLADEKEIAVKRLSKTSSQGVEELKNEIILIAKLQHRNLVRLLACCIEQ 180
Query: 409 DEKILVYEYMPNSSLDFHLFNEEKHKHLDWKLRLSIINGIARGLLYLHEDSRLRVIHRDL 468
+EK+L+YEY+PNSSLDFHLF+ K L W+ RL+IINGIA+GLLYLHEDSRLRVIHRDL
Sbjct: 181 NEKLLIYEYLPNSSLDFHLFDMVKGAQLAWRQRLNIINGIAKGLLYLHEDSRLRVIHRDL 360
Query: 469 KASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAPEYAMAGLFSVKSDVF 528
KASN+LLD EMNPKISDFGLARTF DQ + T RV GTYGYMAPEYAM GLFSVKSDVF
Sbjct: 361 KASNILLDQEMNPKISDFGLARTFGGDQDEANTIRVVGTYGYMAPEYAMEGLFSVKSDVF 540
Query: 529 SFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWKLWCEGKCLELIDPFHQKT 579
SFGVL+LEII G++N F+LSEH QSL ++ W LWC+ K EL+DP +K+
Sbjct: 541 SFGVLLLEIISGRKNSKFYLSEHGQSLPIFAWNLWCKRKGFELMDPSIEKS 693
>TC79762 similar to PIR|H96731|H96731 hypothetical protein F5A18.8
[imported] - Arabidopsis thaliana, partial (55%)
Length = 1564
Score = 320 bits (821), Expect = 8e-88
Identities = 170/342 (49%), Positives = 225/342 (65%), Gaps = 1/342 (0%)
Frame = +3
Query: 304 LSRTITPISFRNQVQRQDSFNGELPTIPLTIIEQSTDDFSESYKLGEGGFGPVYKGTLPD 363
L+ T + N+ + Q + E + +T +F+ ++KLGEGGFGPVYKG L D
Sbjct: 270 LNAVTTQVEGDNEAELQKMASREQKIFSYETLLSATKNFNATHKLGEGGFGPVYKGKLSD 449
Query: 364 GREVAVKRLSETSSQGSEEFKNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSL 423
GREVAVK+LS+TS+QG +EF NE +A++QH+N+ LLGYC+ G EKILVYEY+P+ SL
Sbjct: 450 GREVAVKKLSQTSNQGKKEFMNEAKLLARVQHKNVVNLLGYCVHGTEKILVYEYVPHESL 629
Query: 424 DFHLFNE-EKHKHLDWKLRLSIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPK 482
D LF E EK + LDWK R II G+A+GLLYLHEDS +IHRD+KASN+LLDD+ K
Sbjct: 630 DKFLFKEAEKREQLDWKRRFGIITGVAKGLLYLHEDSHNCIIHRDIKASNILLDDKWTAK 809
Query: 483 ISDFGLARTFDKDQCQTKTKRVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKR 542
I+DFG+AR F +DQ Q KT RV GT GYMAPEY M G SVK+DVFS+GVLVLE+I G+R
Sbjct: 810 IADFGMARLFPEDQSQVKT-RVAGTNGYMAPEYMMHGRLSVKADVFSYGVLVLELITGQR 986
Query: 543 NGDFFLSEHMQSLLLYTWKLWCEGKCLELIDPFHQKTYIESEVLKCIHIGLLCVQEDAAD 602
N F L +LL + +K++ +G+ LE++D T + +V CI + LLC+Q D
Sbjct: 987 NSSFNLXVEEHNLLDWAYKMYKKGRSLEIVDSALASTVLTEQVDMCIQLALLCIQGDPQL 1166
Query: 603 RPTMSTVVRMLGSDTVDLPKPTQPAFSVGRKSKNEDQISKNS 644
RPTM +V L + PT+P + + QI K S
Sbjct: 1167RPTMRRIVVKLSRKS-----PTKPHGTTNKTRNTR*QI*KTS 1277
>TC88392 similar to GP|11275527|dbj|BAB18292. putative receptor-like protein
kinase {Oryza sativa (japonica cultivar-group)}, partial
(61%)
Length = 1017
Score = 296 bits (759), Expect = 1e-80
Identities = 150/278 (53%), Positives = 194/278 (68%)
Frame = +1
Query: 332 LTIIEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIA 391
L ++ +T+ FSE +LG GGFGPV+KG +P+G EVA+K+LS S QG EF NEV +
Sbjct: 187 LNTLQLATNFFSELNQLGRGGFGPVFKGLMPNGEEVAIKKLSMESRQGIREFTNEVRLLL 366
Query: 392 KLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKHLDWKLRLSIINGIARG 451
++QH+NL LLG C EG EK+LVYEY+PN SLD LF +K + LDW R I+ GIARG
Sbjct: 367 RIQHKNLVTLLGCCAEGPEKMLVYEYLPNKSLDHFLF--DKKRSLDWMTRFRIVTGIARG 540
Query: 452 LLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYM 511
LLYLHE++ R+IHRD+KASN+LLD+++NPKISDFGLAR F + +T R+ T+GYM
Sbjct: 541 LLYLHEEAPERIIHRDIKASNILLDEKLNPKISDFGLARLFPGEDTHVQTFRISRTHGYM 720
Query: 512 APEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWKLWCEGKCLEL 571
APEYA+ G SVK+DVFS+GVLVLEI+ G++N D L LL Y WKL+ K ++L
Sbjct: 721 APEYALRGYLSVKTDVFSYGVLVLEIVSGRKNHDLKLDAEKADLLSYAWKLYQGRKIMDL 900
Query: 572 IDPFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTV 609
ID K Y E CI +GLLC Q +RP M++V
Sbjct: 901 IDQNIGK-YNGDEAAMCIQLGLLCCQASLVERPDMNSV 1011
>TC91639 similar to PIR|T05181|T05181 S-receptor kinase (EC 2.7.1.-)
T6K22.120 precursor - Arabidopsis thaliana, partial
(28%)
Length = 1313
Score = 290 bits (743), Expect = 9e-79
Identities = 158/300 (52%), Positives = 197/300 (65%), Gaps = 16/300 (5%)
Frame = +3
Query: 263 EGTSKAKTLIIIFVS-----ITVAVALLSCWVYSYWRKNRLSKGGML--------SRTIT 309
+G K +++I ++ I + + +L W Y K SK + SR ++
Sbjct: 123 DGGKNEKIMMVIILTSLAGLICIGIIVLLVWRYKRQLKASCSKNSDVLPVFDAHKSREMS 302
Query: 310 ---PISFRNQVQRQDSFNGELPTIPLTIIEQSTDDFSESYKLGEGGFGPVYKGTLPDGRE 366
P S ++ ELP + + +T++FSE KLG+GGFGPVYKG LP G E
Sbjct: 303 AEIPGSVELGLEGNQLSKVELPFFNFSCMSSATNNFSEENKLGQGGFGPVYKGKLPSGEE 482
Query: 367 VAVKRLSETSSQGSEEFKNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFH 426
+AVKRLS S QG +EFKNE+ A+LQHRNL KL+G IEGDEK+LVYE+M N SLD
Sbjct: 483 IAVKRLSRRSGQGLDEFKNEMRLFAQLQHRNLVKLMGCSIEGDEKLLVYEFMLNKSLDRF 662
Query: 427 LFNEEKHKHLDWKLRLSIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDF 486
LF+ K LDW R II GIARGLLYLH DSRLR+IHRDLKASN+LLD+ MNPKISDF
Sbjct: 663 LFDPIKKTQLDWARRYEIIEGIARGLLYLHRDSRLRIIHRDLKASNILLDENMNPKISDF 842
Query: 487 GLARTFDKDQCQTKTKRVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDF 546
GLAR F +Q + +V GTYGYM+PEYAM GL SVKSDV+SFGVL+LEI+ G+RN F
Sbjct: 843 GLARIFGGNQNEENATKVVGTYGYMSPEYAMEGLVSVKSDVYSFGVLLLEIVSGRRNTSF 1022
>BG453591 similar to PIR|T05181|T05 S-receptor kinase (EC 2.7.1.-) T6K22.120
precursor - Arabidopsis thaliana, partial (24%)
Length = 640
Score = 280 bits (717), Expect = 1e-75
Identities = 140/212 (66%), Positives = 169/212 (79%), Gaps = 1/212 (0%)
Frame = +3
Query: 320 QDSFNGELPTIPLTIIEQSTDDFSESYKLGEGGFGPVYKGTLP-DGREVAVKRLSETSSQ 378
+D + ELP L+ I +T+DFS KLGEGGFGPVYKGTL D RE+AVKRLS +S Q
Sbjct: 3 EDEQDFELPFFNLSTIIDATNDFSNDNKLGEGGFGPVYKGTLVLDRREIAVKRLSGSSKQ 182
Query: 379 GSEEFKNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKHLDW 438
G+ EFKNEVI +KLQHRNL K+LG CI+G+EK+L+YEYMPN SLD LF++ + K LDW
Sbjct: 183 GTREFKNEVILCSKLQHRNLVKVLGCCIQGEEKMLIYEYMPNRSLDSFLFDQAQKKLLDW 362
Query: 439 KLRLSIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQ 498
R +II GIARGL+YLH+DSRLR+IHRDLK SN+LLD++MNPKISDFGLA+ DQ +
Sbjct: 363 SKRFNIICGIARGLIYLHQDSRLRIIHRDLKPSNILLDNDMNPKISDFGLAKICGDDQVE 542
Query: 499 TKTKRVFGTYGYMAPEYAMAGLFSVKSDVFSF 530
T RV GT+GYMAPEYA+ GLFS+KSDVFSF
Sbjct: 543 GNTNRVVGTHGYMAPEYAIDGLFSIKSDVFSF 638
>TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.24
[imported] - Arabidopsis thaliana, partial (32%)
Length = 1547
Score = 275 bits (704), Expect = 3e-74
Identities = 148/316 (46%), Positives = 205/316 (64%), Gaps = 2/316 (0%)
Frame = +1
Query: 335 IEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQ 394
I+ +T++F K+GEGGFGPVYKG L DG +AVK+LS S QG+ EF NE+ I+ LQ
Sbjct: 172 IKVATNNFDPKNKIGEGGFGPVYKGVLSDGAVIAVKQLSSKSKQGNREFVNEIGMISALQ 351
Query: 395 HRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNE-EKHKHLDWKLRLSIINGIARGLL 453
H NL KL G CIEG++ +LVYEYM N+SL LF + E+ +LDW+ R+ I GIARGL
Sbjct: 352 HPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGKPEQRLNLDWRTRMKICVGIARGLA 531
Query: 454 YLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAP 513
YLHE+SRL+++HRD+KA+NVLLD +N KISDFGLA+ +++ T R+ GT GYMAP
Sbjct: 532 YLHEESRLKIVHRDIKATNVLLDKNLNAKISDFGLAKLDEEENTHIST-RIAGTIGYMAP 708
Query: 514 EYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWKLWCEGKCLELID 573
EYAM G + K+DV+SFGV+ LEI+ G N ++ E LL + + L +G LEL+D
Sbjct: 709 EYAMRGYLTDKADVYSFGVVALEIVSGMSNTNYRPKEEFVYLLDWAYVLQEQGNLLELVD 888
Query: 574 PFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRML-GSDTVDLPKPTQPAFSVGR 632
P Y E ++ + + LLC RP MS+VV ML G+ + P + + G
Sbjct: 889 PTLGSKYSSEEAMRMLQLALLCTNPSPTLRPPMSSVVSMLEGNTPIQAPIIKRSDSTAGA 1068
Query: 633 KSKNEDQISKNSKDNS 648
+ K + +S++S+ S
Sbjct: 1069RFKAFELLSQDSQTTS 1116
>BG586735 similar to GP|11275527|db putative receptor-like protein kinase
{Oryza sativa (japonica cultivar-group)}, partial (57%)
Length = 794
Score = 175 bits (443), Expect(2) = 7e-72
Identities = 84/147 (57%), Positives = 109/147 (74%)
Frame = +3
Query: 358 KGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEY 417
+G +P+G EVA+K+LS S QG EF NEV + ++QH+NL LLG C EG EK+LVYEY
Sbjct: 6 RGLMPNGEEVAIKKLSMESRQGIREFTNEVRLLLRIQHKNLVTLLGCCAEGPEKMLVYEY 185
Query: 418 MPNSSLDFHLFNEEKHKHLDWKLRLSIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDD 477
+PN SLD LF +K + LDW R I+ GIARGLLYLHE++ R+IHRD+KASN+LLD+
Sbjct: 186 LPNKSLDHFLF--DKKRSLDWMTRFRIVTGIARGLLYLHEEAPERIIHRDIKASNILLDE 359
Query: 478 EMNPKISDFGLARTFDKDQCQTKTKRV 504
++NPKISDFGLAR F + +T R+
Sbjct: 360 KLNPKISDFGLARLFPGEDTHVQTFRI 440
Score = 114 bits (286), Expect(2) = 7e-72
Identities = 60/116 (51%), Positives = 75/116 (63%)
Frame = +2
Query: 509 GYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWKLWCEGKC 568
GYMAPEYA+ G SVK+DVFS+GVLVLEI+ G++N D L LL Y WKL+ GK
Sbjct: 446 GYMAPEYALRGYLSVKTDVFSYGVLVLEIVSGRKNHDLKLDAEKADLLSYAWKLYQGGKI 625
Query: 569 LELIDPFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRMLGSDTVDLPKPT 624
++LID K Y E CI +GLLC Q +RP M++V ML SD+ LPKP+
Sbjct: 626 MDLIDQNIGK-YNGDEAAMCIQLGLLCCQASLVERPDMNSVNLMLSSDSFTLPKPS 790
>BQ136505 similar to PIR|T05754|T05 S-receptor kinase (EC 2.7.1.-) M4I22.110
precursor - Arabidopsis thaliana, partial (23%)
Length = 618
Score = 264 bits (674), Expect = 9e-71
Identities = 130/199 (65%), Positives = 158/199 (79%)
Frame = +2
Query: 364 GREVAVKRLSETSSQGSEEFKNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSL 423
G+EVA+KR S+ S QG EEFKNEV+ IAKLQHRNL KLLG CI +EK+L+YEYMPN SL
Sbjct: 20 GKEVAIKRNSKMSDQGLEEFKNEVLLIAKLQHRNLVKLLGCCIHREEKLLIYEYMPNRSL 199
Query: 424 DFHLFNEEKHKHLDWKLRLSIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKI 483
D+ +F+E + K LDW R II G+ARGLLYLH+DSRLR+IHRDLK SN+LLD MNPKI
Sbjct: 200 DYFIFDETRSKLLDWSKRSHIIAGVARGLLYLHQDSRLRIIHRDLKLSNILLDALMNPKI 379
Query: 484 SDFGLARTFDKDQCQTKTKRVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRN 543
SDFGLARTF DQ + KT+++ GTYGYM PEYA+ G +S+KSDVFSFGV+VLEII GK+
Sbjct: 380 SDFGLARTFCGDQVEAKTRKLVGTYGYMPPEYAVHGRYSMKSDVFSFGVIVLEIISGKKI 559
Query: 544 GDFFLSEHMQSLLLYTWKL 562
F+ H +LL + W+L
Sbjct: 560 KVFYDPXHSLNLLGHAWRL 616
>TC88193 similar to PIR|G96602|G96602 probable receptor protein kinase
F14G9.24 [imported] - Arabidopsis thaliana, partial
(20%)
Length = 1815
Score = 254 bits (648), Expect(2) = 2e-69
Identities = 141/286 (49%), Positives = 189/286 (65%), Gaps = 5/286 (1%)
Frame = +3
Query: 314 RNQVQRQDSFNGELPTIPLTI----IEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAV 369
R ++ D + T+P T ++ +T DF+ KLGEGGFGPVYKGTL DGR VAV
Sbjct: 480 RRKLYNDDDDLVGIDTMPNTFSYYELKNATSDFNRDNKLGEGGFGPVYKGTLNDGRFVAV 659
Query: 370 KRLSETSSQGSEEFKNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFN 429
K+LS S QG +F E+ I+ +QHRNL KL G CIEG++++LVYEY+ N SLD LF
Sbjct: 660 KQLSIGSHQGKSQFIAEIATISAVQHRNLVKLYGCCIEGNKRLLVYEYLENKSLDQALFG 839
Query: 430 EEKHKHLDWKLRLSIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLA 489
L+W R + G+ARGL YLHE+SRLR++HRD+KASN+LLD E+ PK+SDFGLA
Sbjct: 840 NV--LFLNWSTRYDVCMGVARGLTYLHEESRLRIVHRDVKASNILLDSELVPKLSDFGLA 1013
Query: 490 RTFDKDQCQTKTKRVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLS 549
+ +D + T RV GT GY+APEYAM G + K+DVFSFGV+ LE++ G+ N D L
Sbjct: 1014KLYDDKKTHIST-RVAGTIGYLAPEYAMRGRLTEKADVFSFGVVALELVSGRPNSDSSLE 1190
Query: 550 EHMQSLLLYTWKLWCEGKCL-ELIDPFHQKTYIESEVLKCIHIGLL 594
E LL + W+L E C+ +LIDP + +E EV + + IG++
Sbjct: 1191EDKMYLLDWAWQLH-ERNCINDLIDPRLSEFNME-EVERLVGIGIV 1322
Score = 27.7 bits (60), Expect(2) = 2e-69
Identities = 18/59 (30%), Positives = 26/59 (43%)
Frame = +1
Query: 592 GLLCVQEDAADRPTMSTVVRMLGSDTVDLPKPTQPAFSVGRKSKNEDQISKNSKDNSVD 650
GLLC Q RP+MS VV ML D ++P + K + I ++ +D
Sbjct: 1315 GLLCTQTSPNLRPSMSRVVAMLLGDIEVSTVTSRPEYWTDWKFGDVSSIMTDTSAEGLD 1491
>TC92541 similar to PIR|T02153|T02153 protein kinase homolog T1F15.1 -
Arabidopsis thaliana, partial (22%)
Length = 791
Score = 153 bits (386), Expect(3) = 5e-67
Identities = 72/116 (62%), Positives = 92/116 (79%)
Frame = +2
Query: 337 QSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQHR 396
++T DFS KLG+GG+GPVYKG LP G+E+AVKRLS+TS QG EFKNE++ I +LQH
Sbjct: 5 EATIDFSPENKLGQGGYGPVYKGILPTGQEIAVKRLSKTSRQGIVEFKNELVLICELQHT 184
Query: 397 NLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKHLDWKLRLSIINGIARGL 452
NL +LLG CI +E+IL+YEYM N SLDF+LF+ + K LDWK RL+II GI++GL
Sbjct: 185 NLVQLLGCCIHEEERILIYEYMSNKSLDFYLFDSTRRKCLDWKKRLNIIEGISQGL 352
Score = 80.5 bits (197), Expect(3) = 5e-67
Identities = 43/112 (38%), Positives = 62/112 (54%), Gaps = 25/112 (22%)
Frame = +1
Query: 486 FGLARTFDKDQCQTKTKRVFGT-------------------------YGYMAPEYAMAGL 520
FG+AR F + + T R+ GT GYM+PEYAM G+
Sbjct: 454 FGMARMFTQQESVVNTNRIVGT**VL*TFLMIT*RKRF*FFK*FFLSSGYMSPEYAMEGI 633
Query: 521 FSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWKLWCEGKCLELI 572
S KSDV+SFGVL+LEII G+RN F+ + +L+ + W+LW +G+ L+L+
Sbjct: 634 CSTKSDVYSFGVLLLEIICGRRNNSFYDVDRPLNLIGHAWELWNDGEYLQLM 789
Score = 61.2 bits (147), Expect(3) = 5e-67
Identities = 27/34 (79%), Positives = 32/34 (93%)
Frame = +3
Query: 453 LYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDF 486
LYLH+ SRL++IHRDLKASN+LLD+ MNPKISDF
Sbjct: 354 LYLHKYSRLKIIHRDLKASNILLDENMNPKISDF 455
>TC89703 similar to GP|11994595|dbj|BAB02650. receptor-like serine/threonine
kinase {Arabidopsis thaliana}, partial (26%)
Length = 1212
Score = 170 bits (431), Expect(2) = 3e-65
Identities = 84/146 (57%), Positives = 107/146 (72%), Gaps = 1/146 (0%)
Frame = +2
Query: 338 STDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQHRN 397
+T++F S K+GEGGFGPVYKG L DG +AVK LS S QG+ EF NE+ I+ LQH
Sbjct: 20 ATNNFDISNKIGEGGFGPVYKGRLSDGTLIAVKLLSSKSKQGNREFLNEIGMISALQHPQ 199
Query: 398 LAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHK-HLDWKLRLSIINGIARGLLYLH 456
L KL G C+EGD+ +L+YEY+ N+SL LF +H+ LDW R I GIARGL YLH
Sbjct: 200 LVKLYGCCVEGDQLMLIYEYLENNSLARALFGPAEHQIRLDWPTRYKICVGIARGLAYLH 379
Query: 457 EDSRLRVIHRDLKASNVLLDDEMNPK 482
E+SRL+V+HRD+KA+NVLLD ++NPK
Sbjct: 380 EESRLKVVHRDIKATNVLLDKDLNPK 457
Score = 97.4 bits (241), Expect(2) = 3e-65
Identities = 55/132 (41%), Positives = 79/132 (59%)
Frame = +3
Query: 482 KISDFGLARTFDKDQCQTKTKRVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGK 541
KISDFGLA+ +++ T R+ GTYGYMAPEYAM G + K+DV+SFG++ LEI++G
Sbjct: 456 KISDFGLAKLDEEENTHIST-RIAGTYGYMAPEYAMHGYLTDKADVYSFGIVALEILHGS 632
Query: 542 RNGDFFLSEHMQSLLLYTWKLWCEGKCLELIDPFHQKTYIESEVLKCIHIGLLCVQEDAA 601
N E LL + L +G +EL+D + + E + I++ LLC ++
Sbjct: 633 NNTILRQKEEAFHLLDWAHILKEKGNEIELVDKRLGSNFNKEEAMLMINVALLCTNVTSS 812
Query: 602 DRPTMSTVVRML 613
RP MS+VV ML
Sbjct: 813 LRPAMSSVVSML 848
>TC80500 similar to PIR|G96602|G96602 probable receptor protein kinase
F14G9.24 [imported] - Arabidopsis thaliana, partial
(13%)
Length = 1015
Score = 240 bits (612), Expect = 1e-63
Identities = 131/259 (50%), Positives = 167/259 (63%)
Frame = +1
Query: 358 KGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEY 417
+G L DGR+VAVK+LS S QG +F E+ I+ +QHRNL KL G CIEG +++LVYEY
Sbjct: 19 QGILNDGRDVAVKQLSIGSHQGKSQFVAEIATISAVQHRNLVKLYGCCIEGSKRLLVYEY 198
Query: 418 MPNSSLDFHLFNEEKHKHLDWKLRLSIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDD 477
+ N SLD LF L+W R I G+ARGL YLHE+SRLR++HRD+KASN+LLD
Sbjct: 199 LENKSLDQALFGNVLF--LNWSTRYDICMGVARGLTYLHEESRLRIVHRDVKASNILLDS 372
Query: 478 EMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEI 537
E+ PKISDFGLA+ +D + T RV GT GY+APEYAM G + K+DVFSFGV+ LE+
Sbjct: 373 ELVPKISDFGLAKLYDDKKTHIST-RVAGTIGYLAPEYAMRGHLTEKADVFSFGVVALEL 549
Query: 538 IYGKRNGDFFLSEHMQSLLLYTWKLWCEGKCLELIDPFHQKTYIESEVLKCIHIGLLCVQ 597
+ G+ N D L LL + W+L ELIDP + + EV + + I LLC Q
Sbjct: 550 VSGRPNSDSTLEGEKMYLLEWAWQLHERNTINELIDP-RLSEFNKEEVQRLVGIALLCTQ 726
Query: 598 EDAADRPTMSTVVRMLGSD 616
RP+MS VV ML D
Sbjct: 727 TSPTLRPSMSRVVAMLSGD 783
>TC78270 similar to GP|16649103|gb|AAL24403.1 Unknown protein {Arabidopsis
thaliana}, partial (80%)
Length = 1463
Score = 236 bits (601), Expect = 3e-62
Identities = 128/291 (43%), Positives = 183/291 (61%), Gaps = 2/291 (0%)
Frame = +3
Query: 338 STDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQHRN 397
++D+FS + K+GEGGFG VYKG L G+ A+K LS S QG +EF E+ I++++H N
Sbjct: 297 ASDNFSPANKIGEGGFGSVYKGVLKGGKLAAIKVLSTESKQGVKEFLTEINVISEIKHEN 476
Query: 398 LAKLLGYCIEGDEKILVYEYMPNSSLDFHLF-NEEKHKHLDWKLRLSIINGIARGLLYLH 456
L L G C+EGD +ILVY Y+ N+SL L + + DW+ R I G+ARGL +LH
Sbjct: 477 LVILYGCCVEGDHRILVYNYLENNSLSQTLLAGGHSNIYFDWQTRRRICLGVARGLAFLH 656
Query: 457 EDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAPEYA 516
E+ ++HRD+KASN+LLD ++ PKISDFGLA+ T RV GT GY+APEYA
Sbjct: 657 EEVLPHIVHRDIKASNILLDKDLTPKISDFGLAKLIPSYMTHVST-RVAGTIGYLAPEYA 833
Query: 517 MAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWKLWCEGKCLELIDPFH 576
+ G + K+D++SFGVL++EI+ G+ N + L Q +L TW+L+ + +L+D
Sbjct: 834 IRGQLTRKADIYSFGVLLVEIVSGRSNTNTRLPIADQYILETTWQLYERKELAQLVDISL 1013
Query: 577 QKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRML-GSDTVDLPKPTQP 626
+ E K + I LLC Q+ RPTMS+VV+ML G ++ K T+P
Sbjct: 1014NGEFDAEEACKILKIALLCTQDTPKLRPTMSSVVKMLTGEMDINETKITKP 1166
>TC92229 similar to PIR|B86369|B86369 hypothetical protein AAC98010.1
[imported] - Arabidopsis thaliana, partial (44%)
Length = 1347
Score = 233 bits (595), Expect = 1e-61
Identities = 127/288 (44%), Positives = 176/288 (61%), Gaps = 7/288 (2%)
Frame = +2
Query: 335 IEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQ 394
I + T+ FS +GEGGFG VYK +PDGR A+K L S QG EF+ EV I+++
Sbjct: 173 ILEITNGFSSENVIGEGGFGRVYKALMPDGRVGALKLLKAGSGQGEREFRAEVDTISRVH 352
Query: 395 HRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKHLDWKLRLSIINGIARGLLY 454
HR+L L+GYCI +++L+YE++PN +LD HL +E + LDW R+ I G ARGL Y
Sbjct: 353 HRHLVSLIGYCIAEQQRVLIYEFVPNGNLDQHL-HESQWNVLDWPKRMKIAIGAARGLAY 529
Query: 455 LHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAPE 514
LHE ++IHRD+K+SN+LLDD +++DFGLAR D T RV GT+GYMAPE
Sbjct: 530 LHEGCNPKIIHRDIKSSNILLDDSYEAQVADFGLARLTDDTNTHVST-RVMGTFGYMAPE 706
Query: 515 YAMAGLFSVKSDVFSFGVLVLEIIYGKRN-------GDFFLSEHMQSLLLYTWKLWCEGK 567
YA +G + +SDVFSFGV++LE++ G++ GD L E + +LL + G
Sbjct: 707 YATSGKLTDRSDVFSFGVVLLELVTGRKPVDPTQPVGDESLVEWARPILLRAIE---TGD 877
Query: 568 CLELIDPFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRMLGS 615
EL DP + YI+SE+ + I C++ A RP M + R L S
Sbjct: 878 FSELADPRLHRQYIDSEMFRMIEAAAACIRHSAPKRPRMVQIARALDS 1021
>TC79033 similar to GP|21554229|gb|AAM63304.1 somatic embryogenesis
receptor-like kinase putative {Arabidopsis thaliana},
partial (77%)
Length = 1762
Score = 228 bits (581), Expect = 6e-60
Identities = 122/284 (42%), Positives = 174/284 (60%), Gaps = 1/284 (0%)
Frame = +1
Query: 335 IEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQ 394
+ +T++F+ KLGEGGFG VY G L DG ++AVKRL S++ EF EV +A+++
Sbjct: 442 LHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKADMEFAVEVEILARVR 621
Query: 395 HRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKH-LDWKLRLSIINGIARGLL 453
H+NL L GYC EG E+++VY+YMPN SL HL + + LDW R++I G A G++
Sbjct: 622 HKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSTESLLDWNRRMNIAIGSAEGIV 801
Query: 454 YLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAP 513
YLH + +IHRD+KASNVLLD + +++DFG A+ D T RV GT GY+AP
Sbjct: 802 YLHVQATPHIIHRDVKASNVLLDSDFQARVADFGFAKLI-PDGATHVTTRVKGTLGYLAP 978
Query: 514 EYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWKLWCEGKCLELID 573
EYAM G + DV+SFG+L+LE+ GK+ + S +++ + L CE K EL D
Sbjct: 979 EYAMLGKANESCDVYSFGILLLELASGKKPLEKLSSSVKRAINDWALPLACEKKFSELAD 1158
Query: 574 PFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRMLGSDT 617
P Y+E E+ + I + L+C Q RPTM VV +L ++
Sbjct: 1159PRLNGDYVEEELKRVILVALICAQNQPEKRPTMVEVVELLKGES 1290
>BG451315 weakly similar to PIR|D85065|D850 receptor protein kinase-like
protein [imported] - Arabidopsis thaliana, partial (7%)
Length = 653
Score = 226 bits (577), Expect = 2e-59
Identities = 108/219 (49%), Positives = 147/219 (66%)
Frame = +2
Query: 12 FLHFFLFMILTTAQSPFYLYSICENSTEKTLNTSYQSNVNSLLSWINSDSDLGTISNHNI 71
F+ + AQSP Y+ C NSTE++L +Y+SN+N +LS ++SD+ + NH
Sbjct: 2 FISIISLFTKSIAQSPNYVGDDCHNSTEQSLTATYKSNLNKVLSLLSSDAIVSKGYNHTS 181
Query: 72 IGSNNSDDHDNVYGLYGCRGDITGSFCRFCINTAVREIAQRCPNSVSALIWYDVCVMGYT 131
IG N D VYGLY CRGD+TGSFC+FC++TA ++ QRCPN SA+IWY+ C+ Y+
Sbjct: 182 IGENTIDA---VYGLYDCRGDVTGSFCQFCVSTAASDVLQRCPNRASAVIWYNFCIFRYS 352
Query: 132 NQNTTGKVIVTPSWNITGSRNVKDSTELGKAENNMMSLIRKVTTESSPVWATGEFIWSDT 191
N N G + +PSW GS+N+ + EL KAE+NM SLI + T E++ ++A GEF S
Sbjct: 353 NHNFFGNLTTSPSWQRPGSKNITNPQELDKAEDNMQSLISEATLETNKMYAMGEFNLS-I 529
Query: 192 EKRYGLVQCNRDLSKDGCKECLEAMLDLVPQCCGTKVAW 230
EKRYGLVQC DL++ C +CLEAMLD VP+C GTK+ W
Sbjct: 530 EKRYGLVQCXXDLNEKQCNQCLEAMLDKVPKCXGTKIGW 646
>TC78763 somatic embryogenesis receptor kinase 1 [Medicago truncatula]
Length = 2737
Score = 224 bits (572), Expect = 6e-59
Identities = 142/370 (38%), Positives = 196/370 (52%), Gaps = 7/370 (1%)
Frame = +3
Query: 254 PPMPNPGKQEGTSKAKTLIIIFVSITVAVALLSCWVYSYWRKNRLSKGGMLSRTITPISF 313
PP+ PG T + ++ A ++ +++WR+ + P F
Sbjct: 1104 PPISAPGSGGATGAIAGGVAAGAALLFAAPAIA---FAWWRRRK------------PQEF 1238
Query: 314 RNQVQRQDSFN---GELPTIPLTIIEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVK 370
V ++ G+L L ++ +TD FS LG GGFG VYKG L DG VAVK
Sbjct: 1239 FFDVPAEEDPEVHLGQLKRFSLRELQVATDTFSNKNILGRGGFGKVYKGRLADGSLVAVK 1418
Query: 371 RLSETSSQGSE-EFKNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFN 429
RL E + G E +F+ EV I+ HRNL +L G+C+ E++LVY YM N S+ L
Sbjct: 1419 RLKEERTPGGELQFQTEVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRE 1598
Query: 430 EEKHKH-LDWKLRLSIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGL 488
H+ LDW R I G ARGL YLH+ ++IHRD+KA+N+LLD+E + DFGL
Sbjct: 1599 RPPHQEPLDWPTRKRIALGSARGLSYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGL 1778
Query: 489 ARTFDKDQCQTKTKRVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFL 548
A+ D T V GT G++APEY G S K+DVF +G+++LE+I G+R D
Sbjct: 1779 AKLMDYKDTHV-TTAVRGTIGHIAPEYLSTGKSSEKTDVFGYGIMLLELITGQRAFDLAR 1955
Query: 549 SEHMQSLLLYTW--KLWCEGKCLELIDPFHQKTYIESEVLKCIHIGLLCVQEDAADRPTM 606
+ ++L W L E K L+DP + YIE+EV + I + LLC Q DRP M
Sbjct: 1956 LANDDDVMLLDWVKGLLKEKKLEMLVDPDLKTNYIEAEVEQLIQVALLCTQGSPMDRPKM 2135
Query: 607 STVVRMLGSD 616
S VVRML D
Sbjct: 2136 SDVVRMLEGD 2165
>TC87048 similar to GP|8778594|gb|AAF79602.1| F5M15.3 {Arabidopsis
thaliana}, partial (77%)
Length = 1571
Score = 223 bits (567), Expect = 2e-58
Identities = 124/290 (42%), Positives = 180/290 (61%), Gaps = 5/290 (1%)
Frame = +1
Query: 338 STDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQHRN 397
+T F E+ +GEGGFG V+KG L G VAVK+LS QG +EF EV+ ++ L H N
Sbjct: 454 ATRGFKEANLIGEGGFGKVFKGRLSTGELVAVKQLSHDGRQGFQEFVTEVLMLSLLHHSN 633
Query: 398 LAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKH-LDWKLRLSIINGIARGLLYLH 456
L KL+GYC +GD+++LVYEYMP SL+ HLF+ + K L W R+ I G ARGL YLH
Sbjct: 634 LVKLIGYCTDGDQRLLVYEYMPMGSLEDHLFDLPQDKEPLSWSSRMKIAVGAARGLEYLH 813
Query: 457 EDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAPEYA 516
+ VI+RDLK++N+LLD + +PK+SDFGLA+ + RV GTYGY APEYA
Sbjct: 814 CKADPPVIYRDLKSANILLDSDFSPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYA 993
Query: 517 MAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWKLWCE-GKCLELIDPF 575
M+G ++KSD++SFGV++LE+I G+R D Q+L+ ++ + + K + + DP
Sbjct: 994 MSGKLTLKSDIYSFGVVLLELITGRRAIDASKKPGEQNLVSWSRPYFSDRRKFVHMADPL 1173
Query: 576 HQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVV---RMLGSDTVDLPK 622
Q + + + I I +C+QE RP + +V L S + ++P+
Sbjct: 1174LQGHFPVRCLHQAIAITAMCLQEQPKFRPLIGDIVVALEYLASQSQNIPE 1323
>TC93437 similar to GP|11227578|emb|CAC16506. unnamed protein product
{Arabidopsis thaliana}, partial (62%)
Length = 978
Score = 219 bits (559), Expect = 2e-57
Identities = 112/238 (47%), Positives = 158/238 (66%), Gaps = 1/238 (0%)
Frame = +3
Query: 335 IEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQ 394
+ +T DF S K+G GG+G VYKG L DG +VA+K LS S QG+ EF E+ I+ +Q
Sbjct: 228 LRSATGDFHPSCKIGGGGYGVVYKGVLRDGTQVAIKSLSVESKQGTHEFMTEIAMISNIQ 407
Query: 395 HRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLF-NEEKHKHLDWKLRLSIINGIARGLL 453
H NL KL+G+CIEG+ +ILVYE++ N+SL L ++ K LDW+ R I G A GL
Sbjct: 408 HPNLVKLIGFCIEGNHRILVYEFLENNSLTSSLLGSKSKCVPLDWQKRAIICRGTASGLS 587
Query: 454 YLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAP 513
+LHE+++ ++HRD+KASN+LLD+ +PKI DFGLA+ F + T RV GT GY+AP
Sbjct: 588 FLHEEAQPNIVHRDIKASNILLDENFHPKIGDFGLAKLFPDNVTHVST-RVAGTMGYLAP 764
Query: 514 EYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWKLWCEGKCLEL 571
EYA+ + K+DV+SFG+L+LEII GK + +++ L+ + WKL + LEL
Sbjct: 765 EYALLRQLTKKADVYSFGILMLEIISGKSSXKAAFGDNILVLVEWAWKLKEXNRLLEL 938
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.136 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,608,033
Number of Sequences: 36976
Number of extensions: 362038
Number of successful extensions: 3522
Number of sequences better than 10.0: 738
Number of HSP's better than 10.0 without gapping: 2817
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2886
length of query: 659
length of database: 9,014,727
effective HSP length: 102
effective length of query: 557
effective length of database: 5,243,175
effective search space: 2920448475
effective search space used: 2920448475
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147431.3


