

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147364.1 - phase: 0
(200 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
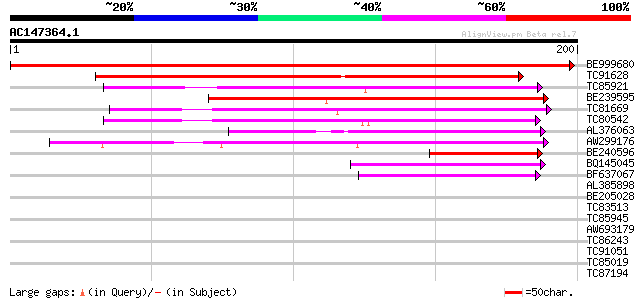
Score E
Sequences producing significant alignments: (bits) Value
BE999680 weakly similar to PIR|B71421|B714 hypothetical protein ... 390 e-109
TC91628 weakly similar to PIR|B71421|B71421 hypothetical protein... 187 3e-48
TC85921 weakly similar to GP|11908074|gb|AAG41466.1 unknown prot... 99 1e-21
BE239595 weakly similar to GP|21592507|gb| unknown {Arabidopsis ... 93 6e-20
TC81669 weakly similar to GP|21739230|emb|CAD40985. OSJNBa0072F1... 82 1e-16
TC80542 similar to GP|18087595|gb|AAL58928.1 unknown protein {Ar... 65 1e-11
AL376063 weakly similar to PIR|H85044|H85 hypothetical protein A... 64 4e-11
AW299176 weakly similar to GP|12322903|gb unknown protein; 80797... 62 2e-10
BE240596 60 6e-10
BQ145045 weakly similar to PIR|H85044|H850 hypothetical protein ... 47 5e-06
BF637067 weakly similar to GP|21618058|gb| unknown {Arabidopsis ... 44 3e-05
AL385898 30 0.62
BE205028 28 3.1
TC83513 27 4.0
TC85945 similar to GP|17473511|gb|AAL38379.1 AT4g33090/F4I10_20 ... 27 4.0
AW693179 weakly similar to GP|21594014|gb| unknown {Arabidopsis ... 27 4.0
TC86243 similar to GP|13877531|gb|AAK43843.1 Unknown protein {Ar... 27 5.3
TC91051 similar to PIR|H84787|H84787 probable receptor-like prot... 27 5.3
TC85019 weakly similar to GP|9293890|dbj|BAB01793.1 emb|CAB70981... 27 5.3
TC87194 NAC1 27 6.9
>BE999680 weakly similar to PIR|B71421|B714 hypothetical protein -
Arabidopsis thaliana, partial (32%)
Length = 616
Score = 390 bits (1002), Expect = e-109
Identities = 198/199 (99%), Positives = 198/199 (99%)
Frame = +3
Query: 1 MEGRGKAIIDGVEGNYGRDELAMKKPPAGGGSCELVLRFLGFVLTLAAAIVVGTDKQTTI 60
MEGRGKAIIDGVEGNYGRDELAMKKPPAGGGSCELVL FLGFVLTLAAAIVVGTDKQTTI
Sbjct: 18 MEGRGKAIIDGVEGNYGRDELAMKKPPAGGGSCELVLXFLGFVLTLAAAIVVGTDKQTTI 197
Query: 61 VPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVANAIACTYGAISMLLTLLNRGKSKVFWGT 120
VPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVANAIACTYGAISMLLTLLNRGKSKVFWGT
Sbjct: 198 VPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVANAIACTYGAISMLLTLLNRGKSKVFWGT 377
Query: 121 LITIFDALMVALLFSGNGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLG 180
LITIFDALMVALLFSGNGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLG
Sbjct: 378 LITIFDALMVALLFSGNGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLG 557
Query: 181 SLVFLLLVVLLPILRSRRR 199
SLVFLLLVVLLPILRSRRR
Sbjct: 558 SLVFLLLVVLLPILRSRRR 614
>TC91628 weakly similar to PIR|B71421|B71421 hypothetical protein -
Arabidopsis thaliana, partial (30%)
Length = 731
Score = 187 bits (474), Expect = 3e-48
Identities = 91/151 (60%), Positives = 118/151 (77%)
Frame = +2
Query: 31 GSCELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFL 90
G+C +VLR VLTL AA+V+ DKQTT+VP+K+ DSLPPL+V V+AKW Y+SA+VYF+
Sbjct: 281 GTCSMVLRLCALVLTLTAAVVLVADKQTTVVPVKISDSLPPLDVPVTAKWQYVSAYVYFV 460
Query: 91 VANAIACTYGAISMLLTLLNRGKSKVFWGTLITIFDALMVALLFSGNGAATAIGVLGYQG 150
VAN IA Y +S ++ L N KSK+ TL+T+ DA+MVALLFSGNGAA AIGVL +G
Sbjct: 461 VANFIAFAYATLSFVIALANGHKSKLL-VTLVTLLDAIMVALLFSGNGAAWAIGVLAEKG 637
Query: 151 NSHVRWKKVCNVFDKYCHQVAASIILSQLGS 181
NSHV W K+C+VFDK+C+Q AA+ ++S LGS
Sbjct: 638 NSHVMWNKMCHVFDKFCNQAAAACLISLLGS 730
>TC85921 weakly similar to GP|11908074|gb|AAG41466.1 unknown protein
{Arabidopsis thaliana}, partial (38%)
Length = 1032
Score = 98.6 bits (244), Expect = 1e-21
Identities = 54/157 (34%), Positives = 89/157 (56%), Gaps = 2/157 (1%)
Frame = +3
Query: 34 ELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVAN 93
+L+LRFL F ++A+ +V+ T QT +P V AK+ Y AFVYF+ A
Sbjct: 282 DLILRFLLFAASVASVVVMVTGNQTVYLP-----------VPRPAKFRYSPAFVYFVAAF 428
Query: 94 AIACTYGAISMLLTLLNRGKSKVFWGTLITI--FDALMVALLFSGNGAATAIGVLGYQGN 151
++A Y I+ ++L K + L+ + +DA+M+ +L S G A ++ LG +GN
Sbjct: 429 SVAGLYSIITTFISLSAIRKPNLKTKLLLHLIFWDAVMLGILASATGTAGSVAYLGLKGN 608
Query: 152 SHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLV 188
H W K+C+++DK+C + AS+ + GS+V LLL+
Sbjct: 609 KHTDWHKICHIYDKFCRHIGASVGVGLFGSIVTLLLI 719
>BE239595 weakly similar to GP|21592507|gb| unknown {Arabidopsis thaliana},
partial (44%)
Length = 651
Score = 93.2 bits (230), Expect = 6e-20
Identities = 46/121 (38%), Positives = 75/121 (61%), Gaps = 1/121 (0%)
Frame = +1
Query: 71 PLNVAVSAKWHYLSAFVYFLVANAIACTYGAISMLLTLLN-RGKSKVFWGTLITIFDALM 129
P+ V ++AK+ + AFVYF+VAN I + + +L + +K LI +FD +
Sbjct: 163 PITVPLTAKFQHTPAFVYFVVANGIVSLHNLEMIAKHILGPKFHNKGLQLALIAVFDTMA 342
Query: 130 VALLFSGNGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVV 189
+AL SG+GAATA+ LG GNSH +W K+C+ F+ YC++ S+I S +G ++ L++ V
Sbjct: 343 LALASSGDGAATAMSELGRNGNSHAKWNKICDKFESYCNRGGGSLIASFIGLILLLIITV 522
Query: 190 L 190
+
Sbjct: 523 M 525
>TC81669 weakly similar to GP|21739230|emb|CAD40985. OSJNBa0072F16.9 {Oryza
sativa}, partial (15%)
Length = 859
Score = 82.4 bits (202), Expect = 1e-16
Identities = 46/158 (29%), Positives = 82/158 (51%), Gaps = 2/158 (1%)
Frame = +3
Query: 36 VLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVANAI 95
+LR L VLT + +V T+ Q+ ++ ++ A ++Y S+F +F+ AN +
Sbjct: 174 ILRILVIVLTAVSIVVTVTNNQSVML----------FSIRFEAHFYYTSSFKFFVAANGV 323
Query: 96 ACTYGAISMLLTLLNRGKS--KVFWGTLITIFDALMVALLFSGNGAATAIGVLGYQGNSH 153
C ++++ LL R ++ + + L+ + D +M LL +G AATA+G +G G H
Sbjct: 324 VCFMSVLTLIFNLLMRQQTPQRKDYYFLLFLVDLVMTVLLIAGCSAATAVGYVGQYGEKH 503
Query: 154 VRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVVLL 191
V W +C+ K+C S++LS L L L +L+
Sbjct: 504 VGWTAICDHVQKFCKTNLISLLLSYLAFFANLGLTILI 617
>TC80542 similar to GP|18087595|gb|AAL58928.1 unknown protein {Arabidopsis
thaliana}, partial (66%)
Length = 1262
Score = 65.5 bits (158), Expect = 1e-11
Identities = 45/158 (28%), Positives = 75/158 (46%), Gaps = 4/158 (2%)
Frame = +3
Query: 34 ELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVAN 93
ELVLRF+ L + A +++ TD Q + AK+ + A V+ +VAN
Sbjct: 528 ELVLRFVNLGLGVLAVVLIVTDSQVREF----------FTIQKKAKFTDMKALVFLVVAN 677
Query: 94 AIACTYGAISMLLTLLNRGKSKVFWGTLIT--IF--DALMVALLFSGNGAATAIGVLGYQ 149
AIA Y I L +++ K V + + IF D +M + + AA V
Sbjct: 678 AIAAGYSLIQGLRCVVSMIKGSVLFNKPLAWAIFSCDQVMAYITVAAVSAAAQSAVFAKM 857
Query: 150 GNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLL 187
G ++W K+CN++ K+C+QV + + L SL +++
Sbjct: 858 GQEQLQWMKICNMYGKFCNQVGEGLASAFLVSLSMVVV 971
>AL376063 weakly similar to PIR|H85044|H85 hypothetical protein AT4g03540
[imported] - Arabidopsis thaliana, partial (73%)
Length = 504
Score = 63.9 bits (154), Expect = 4e-11
Identities = 32/112 (28%), Positives = 58/112 (51%)
Frame = +1
Query: 78 AKWHYLSAFVYFLVANAIACTYGAISMLLTLLNRGKSKVFWGTLITIFDALMVALLFSGN 137
AK+ AF YF++AN+I YG + L + W L+ D ++ LL S
Sbjct: 4 AKYTNSPAFKYFVIANSIVTVYGFFVLFLPA-----ESLLW-RLVVAMDMVLTMLLISSI 165
Query: 138 GAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVV 189
AA AI + GN++ W +C K+C+ + +++ S +G +++L+L++
Sbjct: 166 SAALAIAQVAKNGNNYAAWLPICGSVPKFCNHMTGALVASFIGVIIYLILLL 321
>AW299176 weakly similar to GP|12322903|gb unknown protein; 80797-81587
{Arabidopsis thaliana}, partial (78%)
Length = 639
Score = 61.6 bits (148), Expect = 2e-10
Identities = 46/185 (24%), Positives = 82/185 (43%), Gaps = 9/185 (4%)
Frame = +1
Query: 15 NYGRDELAMKKPPAGGG------SCELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDS 68
N +++ + P GG + +LR +AAA +GT QT
Sbjct: 31 NINKEKTILVAPARPGGWKKGTAIMDFILRLGAIAAAIAAAASMGTSDQT---------- 180
Query: 69 LPPLN--VAVSAKWHYLSAFVYFLVANAIACTYGAISMLLTLLNRGKSKVFWGTL-ITIF 125
LP A + + F +F+++ AIA +Y +S+ +++ + L + I
Sbjct: 181 LPFFTQFFQFEASYDSFTTFQFFVISMAIAASYLVLSLPFSIVAIIRPHAPGPRLFLIIL 360
Query: 126 DALMVALLFSGNGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFL 185
D + + L S AA +I L + GN W +CN F +C Q + +++ S + +V +
Sbjct: 361 DTVFLTLATSSAAAAASIVYLAHNGNQDTNWLAICNQFGDFCAQTSGAVVASFITVVVLI 540
Query: 186 LLVVL 190
LLVV+
Sbjct: 541 LLVVM 555
>BE240596
Length = 249
Score = 60.1 bits (144), Expect = 6e-10
Identities = 25/40 (62%), Positives = 32/40 (79%)
Frame = +2
Query: 149 QGNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLV 188
+GNSHV+W KVCNVFD YC + +++LS +GS VFLLLV
Sbjct: 5 KGNSHVQWMKVCNVFDAYCRHMTVALVLSIIGSTVFLLLV 124
>BQ145045 weakly similar to PIR|H85044|H850 hypothetical protein AT4g03540
[imported] - Arabidopsis thaliana, partial (46%)
Length = 464
Score = 47.0 bits (110), Expect = 5e-06
Identities = 19/69 (27%), Positives = 39/69 (55%)
Frame = +1
Query: 121 LITIFDALMVALLFSGNGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLG 180
LI + ++ LL S AA AI + GN++ W +C K+C+ + +++ S +G
Sbjct: 118 LIDFSEQVLTMLLISSISAALAIAQVAKNGNNYAAWLPICGSVPKFCNHMTGALVASFIG 297
Query: 181 SLVFLLLVV 189
+++L+L++
Sbjct: 298 VIIYLILLL 324
>BF637067 weakly similar to GP|21618058|gb| unknown {Arabidopsis thaliana},
partial (25%)
Length = 610
Score = 44.3 bits (103), Expect = 3e-05
Identities = 17/64 (26%), Positives = 36/64 (55%)
Frame = +3
Query: 124 IFDALMVALLFSGNGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLV 183
+ D ++V L F+ N +A + G+ +W KVCN F ++C+Q+ +I+ + S++
Sbjct: 279 LLDQIVVYLTFATNTSAFEACLFALTGSEAFQWMKVCNKFTRFCYQIGGAILCCYIASIL 458
Query: 184 FLLL 187
++
Sbjct: 459 MAMI 470
>AL385898
Length = 461
Score = 30.0 bits (66), Expect = 0.62
Identities = 21/70 (30%), Positives = 34/70 (48%), Gaps = 10/70 (14%)
Frame = +1
Query: 116 VFWGTLITIFDALMVALLFSGNGAATAIGVLGYQGNSHV---------RW-KKVCNVFDK 165
V GTL ++AL++ SG G+ + + Y G + RW +KVCN+FD+
Sbjct: 229 VLGGTLWGSWNALILVSFCSGTGSTISYLLSYYLGQPIIQNYLFERMERWNEKVCNIFDE 408
Query: 166 YCHQVAASII 175
+ S+I
Sbjct: 409 LIIYILISLI 438
>BE205028
Length = 463
Score = 27.7 bits (60), Expect = 3.1
Identities = 14/38 (36%), Positives = 24/38 (62%)
Frame = +3
Query: 154 VRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVVLL 191
V W + F +C+ + A+I+ S LG++ +LL+V LL
Sbjct: 69 VSWFSLLLGFFPFCYFLYAAIVASVLGAVFWLLVVRLL 182
>TC83513
Length = 629
Score = 27.3 bits (59), Expect = 4.0
Identities = 15/45 (33%), Positives = 24/45 (53%)
Frame = +2
Query: 83 LSAFVYFLVANAIACTYGAISMLLTLLNRGKSKVFWGTLITIFDA 127
L FV+F+V + C Y L+ L + K+FWG + +FD+
Sbjct: 371 LLLFVFFVVISGGFCLY-----LI*LKFC*RXKIFWGLISIVFDS 490
>TC85945 similar to GP|17473511|gb|AAL38379.1 AT4g33090/F4I10_20
{Arabidopsis thaliana}, partial (94%)
Length = 2944
Score = 27.3 bits (59), Expect = 4.0
Identities = 19/61 (31%), Positives = 29/61 (47%)
Frame = +1
Query: 89 FLVANAIACTYGAISMLLTLLNRGKSKVFWGTLITIFDALMVALLFSGNGAATAIGVLGY 148
F+V NA T ++ T NR SKVF + + +F+ + +L T +GVL
Sbjct: 208 FIVLNAAELTVSDDAVSFT--NRDSSKVFKPSKVELFEDDEILVLEFSEKIPTGLGVLAI 381
Query: 149 Q 149
Q
Sbjct: 382 Q 384
>AW693179 weakly similar to GP|21594014|gb| unknown {Arabidopsis thaliana},
partial (46%)
Length = 409
Score = 27.3 bits (59), Expect = 4.0
Identities = 17/70 (24%), Positives = 28/70 (39%)
Frame = +1
Query: 35 LVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVANA 94
LV R F +LA++I + +T P +W++ F + + +NA
Sbjct: 187 LVFRLTSFSFSLASSIFMLATSRTGDSP----------------RWYHFDTFRFVVASNA 318
Query: 95 IACTYGAISM 104
I TY M
Sbjct: 319 IVATYSLFEM 348
>TC86243 similar to GP|13877531|gb|AAK43843.1 Unknown protein {Arabidopsis
thaliana}, partial (41%)
Length = 841
Score = 26.9 bits (58), Expect = 5.3
Identities = 10/32 (31%), Positives = 18/32 (56%)
Frame = -3
Query: 151 NSHVRWKKVCNVFDKYCHQVAASIILSQLGSL 182
NSH++W K+ V ++C Q + +GS+
Sbjct: 653 NSHIKWNKI--VIQQHCTQALNHLFAGNIGSV 564
>TC91051 similar to PIR|H84787|H84787 probable receptor-like protein kinase
[imported] - Arabidopsis thaliana, partial (36%)
Length = 1240
Score = 26.9 bits (58), Expect = 5.3
Identities = 22/58 (37%), Positives = 29/58 (49%), Gaps = 6/58 (10%)
Frame = +3
Query: 50 IVVGTDKQTTIVPIKV-VDSLPPLNVAVSAKWHYLSAFVYFL-----VANAIACTYGA 101
+ VGT+K +T VPI V D +PP+ V +A + Y L NA A TY A
Sbjct: 714 VAVGTEKVSTNVPILVNRDDVPPVKVMQTAVVGTNGSLTYRLNLDGFPGNAWAVTYFA 887
>TC85019 weakly similar to GP|9293890|dbj|BAB01793.1
emb|CAB70981.1~gene_id:MVE11.3~similar to unknown
protein {Arabidopsis thaliana}, partial (10%)
Length = 663
Score = 26.9 bits (58), Expect = 5.3
Identities = 20/77 (25%), Positives = 33/77 (41%)
Frame = +2
Query: 32 SCELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFLV 91
SC LV + + AA V G + Q +PI + D F+ F+V
Sbjct: 347 SCMLVATLIATITFAAAITVPGGNNQDKGIPIFLSD----------------KTFMLFIV 478
Query: 92 ANAIACTYGAISMLLTL 108
++A+A +S+L+ L
Sbjct: 479 SDALALFSSMVSLLMFL 529
>TC87194 NAC1
Length = 2067
Score = 26.6 bits (57), Expect = 6.9
Identities = 14/42 (33%), Positives = 21/42 (49%), Gaps = 6/42 (14%)
Frame = +1
Query: 83 LSAFVYFLVAN------AIACTYGAISMLLTLLNRGKSKVFW 118
LSAF+Y LV N C GA +++ NR ++ +W
Sbjct: 1909 LSAFIYILVVNK*LK*VMYYCDLGAFPLIIVSENREWTRSWW 2034
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.141 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,672,518
Number of Sequences: 36976
Number of extensions: 91289
Number of successful extensions: 720
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 707
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 713
length of query: 200
length of database: 9,014,727
effective HSP length: 91
effective length of query: 109
effective length of database: 5,649,911
effective search space: 615840299
effective search space used: 615840299
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC147364.1


