

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147363.5 - phase: 0
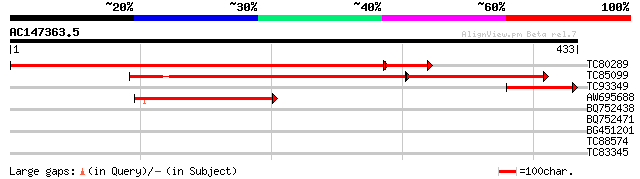
(433 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC80289 similar to GP|13194812|gb|AAK15568.1 putative hydroxymet... 577 e-180
TC85099 similar to GP|13194812|gb|AAK15568.1 putative hydroxymet... 267 e-119
TC93349 similar to GP|13194812|gb|AAK15568.1 putative hydroxymet... 104 6e-23
AW695688 similar to GP|13194812|gb| putative hydroxymethylglutar... 89 3e-18
BQ752438 similar to GP|12044690|e pyruvate carboxylase {Aspergil... 33 0.21
BQ752471 GP|16117372|g ICP4 {Macropodid herpesvirus 1}, partial ... 29 3.1
BG451201 weakly similar to GP|21114152|gb ATP dependent RNA heli... 28 5.3
TC88574 weakly similar to GP|22137236|gb|AAM91463.1 At2g36630/F1... 28 5.3
TC83345 similar to GP|18086395|gb|AAL57656.1 AT5g13210/T31B5_30 ... 28 9.0
>TC80289 similar to GP|13194812|gb|AAK15568.1 putative
hydroxymethylglutaryl-CoA lyase {Arabidopsis thaliana},
partial (48%)
Length = 1178
Score = 577 bits (1488), Expect(2) = e-180
Identities = 285/289 (98%), Positives = 288/289 (99%)
Frame = +2
Query: 1 MSSLEEPLGHDKLPSMSTMDRVQRFSSGCCRPQVDNLGMGNCFIEGRSCSTSNSCNEDNE 60
MSSLEEPLGHDKLPSMSTMDRVQRFSSGCCRPQVDNLGMGNCFIEGRSCSTSNSCNEDNE
Sbjct: 206 MSSLEEPLGHDKLPSMSTMDRVQRFSSGCCRPQVDNLGMGNCFIEGRSCSTSNSCNEDNE 385
Query: 61 DYTAETYPWKRQTRDMSRGDSFSPRTMTTGRNTLKSGIVDNSFYTSDYQYSQKRNNKDMQ 120
DYTAETYPWKRQTRDMSRGDSFSPRTMTTGRNTLKSGIVDNSFYTSDYQYSQKRNNKDMQ
Sbjct: 386 DYTAETYPWKRQTRDMSRGDSFSPRTMTTGRNTLKSGIVDNSFYTSDYQYSQKRNNKDMQ 565
Query: 121 DMAYKFMKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSP 180
DMAYKFMKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSP
Sbjct: 566 DMAYKFMKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSP 745
Query: 181 KWVPQLADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNI 240
KWVPQLADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNI
Sbjct: 746 KWVPQLADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNI 925
Query: 241 NCSIEESLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYV 289
NCSIEESLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSK +++
Sbjct: 926 NCSIEESLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKSSFM 1072
Score = 74.3 bits (181), Expect(2) = e-180
Identities = 34/36 (94%), Positives = 36/36 (99%)
Frame = +1
Query: 288 YVAKALYDMGCFEISLGDTIGVGTPGTVVPMLLAVM 323
YVAKALYDMGCF+ISLGDTIGVGTPGTV+PMLLAVM
Sbjct: 1069 YVAKALYDMGCFQISLGDTIGVGTPGTVIPMLLAVM 1176
>TC85099 similar to GP|13194812|gb|AAK15568.1 putative
hydroxymethylglutaryl-CoA lyase {Arabidopsis thaliana},
partial (60%)
Length = 973
Score = 267 bits (683), Expect(2) = e-119
Identities = 135/215 (62%), Positives = 168/215 (77%)
Frame = +1
Query: 92 NTLKSGIVDNSFYTSDYQYSQKRNNKDMQDMAYKFMKGMPEFVKIVEVGPRDGLQNEKNI 151
N+ K ++++ + + N++ + K +P+FVKIVEVG RDGLQNEK I
Sbjct: 25 NSAKGAVLESKLVAIATYFYRYNNSQ----CSISLRKNIPDFVKIVEVGARDGLQNEKAI 192
Query: 152 VPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADAKDVMQAVHNLRGIRLPVLTPNL 211
VPTDVKIELI L S+GLSV+EATSFVSPKWVPQLADAKDV+ AV ++ G PVLTPNL
Sbjct: 193 VPTDVKIELIKLLVSSGLSVVEATSFVSPKWVPQLADAKDVLAAVRDVEGASFPVLTPNL 372
Query: 212 KGFEAAVAAGAREVAVFASASESFSKSNINCSIEESLSRYRAVTRAAKELSIPVRGYVSC 271
KGFEAAVAAGA+EVAVF +ASESFSK+N+NC IE++L+R R + A++ L+IPVRGY+SC
Sbjct: 373 KGFEAAVAAGAKEVAVFPAASESFSKANLNCGIEDNLARCRDIASASRSLTIPVRGYISC 552
Query: 272 VVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDT 306
VVGCP+EG + P KVAYVAK+LY+MG EISLGDT
Sbjct: 553 VVGCPLEGRIAPVKVAYVAKSLYEMGISEISLGDT 657
Score = 179 bits (454), Expect(2) = e-119
Identities = 87/108 (80%), Positives = 97/108 (89%)
Frame = +2
Query: 304 GDTIGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAG 363
G IGVGTPGTV+PML AV+ +VP +KLAVHFHDTYGQ+L NIL+SLQMGIS DSSV+G
Sbjct: 650 GIPIGVGTPGTVIPMLEAVLDVVPIDKLAVHFHDTYGQALSNILISLQMGISTGDSSVSG 829
Query: 364 LGGCPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQL 411
LGGCPYAKGA+GNVATEDVVYMLNGLG+KTNVD+GKLM AGDFI K L
Sbjct: 830 LGGCPYAKGATGNVATEDVVYMLNGLGVKTNVDLGKLMHAGDFICKHL 973
>TC93349 similar to GP|13194812|gb|AAK15568.1 putative
hydroxymethylglutaryl-CoA lyase {Arabidopsis thaliana},
partial (9%)
Length = 580
Score = 104 bits (260), Expect = 6e-23
Identities = 53/54 (98%), Positives = 53/54 (98%)
Frame = +2
Query: 380 EDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIALNRVTADASKI 433
EDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRP GSKTAIALNRVTADASKI
Sbjct: 2 EDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPFGSKTAIALNRVTADASKI 163
>AW695688 similar to GP|13194812|gb| putative hydroxymethylglutaryl-CoA lyase
{Arabidopsis thaliana}, partial (13%)
Length = 543
Score = 89.4 bits (220), Expect = 3e-18
Identities = 55/113 (48%), Positives = 69/113 (60%), Gaps = 4/113 (3%)
Frame = +1
Query: 96 SGIVDN---SFYTSDYQYSQKRNNKDMQDMAYKFMKGMPEFVKIVEVGPRDGLQNEKNIV 152
SG DN + Y + + N ++ + K K +P+FVKIVEVG RDGLQNEK IV
Sbjct: 187 SGANDNHHAASYVVNRHFRADNNGICTKEFSSKLRKNIPDFVKIVEVGARDGLQNEKAIV 366
Query: 153 PTDVKIELIHRLASTGLSVIEATSFVS-PKWVPQLADAKDVMQAVHNLRGIRL 204
PTDVKIELI L S+GLSV +S +LADAKDV+ AV ++ G RL
Sbjct: 367 PTDVKIELIKLLVSSGLSVCVRRQVLSHQNGCHRLADAKDVLAAVRDVEGARL 525
>BQ752438 similar to GP|12044690|e pyruvate carboxylase {Aspergillus niger},
partial (23%)
Length = 949
Score = 33.1 bits (74), Expect = 0.21
Identities = 17/66 (25%), Positives = 31/66 (46%)
Frame = +1
Query: 301 ISLGDTIGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSS 360
+ + D GV P ++ A+ P + VH HD+ G + +++ + G AVD++
Sbjct: 58 LGIKDMAGVLKPHAATLLIGAIRKKYPDLPIHVHTHDSAGTGVASMVACAKAGADAVDAA 237
Query: 361 VAGLGG 366
L G
Sbjct: 238 TDSLSG 255
>BQ752471 GP|16117372|g ICP4 {Macropodid herpesvirus 1}, partial (1%)
Length = 567
Score = 29.3 bits (64), Expect = 3.1
Identities = 14/48 (29%), Positives = 27/48 (56%)
Frame = +1
Query: 83 SPRTMTTGRNTLKSGIVDNSFYTSDYQYSQKRNNKDMQDMAYKFMKGM 130
SPR + T + G++ FYTS + S+ ++ + +QD +K + G+
Sbjct: 172 SPRQLRTLLIGVLIGVIFVLFYTSSLRSSEAKDERTIQDFYHKTVNGL 315
>BG451201 weakly similar to GP|21114152|gb ATP dependent RNA helicase
{Xanthomonas campestris pv. campestris str. ATCC 33913},
partial (4%)
Length = 449
Score = 28.5 bits (62), Expect = 5.3
Identities = 19/45 (42%), Positives = 23/45 (50%)
Frame = -1
Query: 46 GRSCSTSNSCNEDNEDYTAETYPWKRQTRDMSRGDSFSPRTMTTG 90
GRSCSTS + + + + P R TR S S SPRT TG
Sbjct: 305 GRSCSTSRAISRPSC*GSTSCVPTPRATRASS--PS*SPRTSATG 177
>TC88574 weakly similar to GP|22137236|gb|AAM91463.1 At2g36630/F1O11.26
{Arabidopsis thaliana}, partial (25%)
Length = 663
Score = 28.5 bits (62), Expect = 5.3
Identities = 16/63 (25%), Positives = 28/63 (44%)
Frame = +3
Query: 296 MGCFEISLGDTIGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGIS 355
+G F S G GVG G VPML+ ++ A+ G ++ + +L++
Sbjct: 279 IGIFGASFGSVGGVGGGGIFVPMLILIIGFDAKSATAISKCMVTGAAISTVFFNLKLRHP 458
Query: 356 AVD 358
+D
Sbjct: 459 TLD 467
>TC83345 similar to GP|18086395|gb|AAL57656.1 AT5g13210/T31B5_30
{Arabidopsis thaliana}, partial (45%)
Length = 1085
Score = 27.7 bits (60), Expect = 9.0
Identities = 21/82 (25%), Positives = 34/82 (40%), Gaps = 17/82 (20%)
Frame = +1
Query: 230 SASESFSKSNINCS--------------IEESLSRYRAVTRAAKELSIPVRGYV---SCV 272
S SF +S + C +EE+ YR R K++ +P+R +
Sbjct: 223 SVDSSFDRSTLLCETIAKKIFPREEYEGVEEAHYAYRVRDRLRKDVLVPLRKVLELPEVF 402
Query: 273 VGCPVEGPVPPSKVAYVAKALY 294
+G G +P ++VA VA Y
Sbjct: 403 IGANQWGLIPYNRVASVAMKFY 468
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.133 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,737,039
Number of Sequences: 36976
Number of extensions: 153674
Number of successful extensions: 720
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 715
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 719
length of query: 433
length of database: 9,014,727
effective HSP length: 99
effective length of query: 334
effective length of database: 5,354,103
effective search space: 1788270402
effective search space used: 1788270402
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147363.5


