

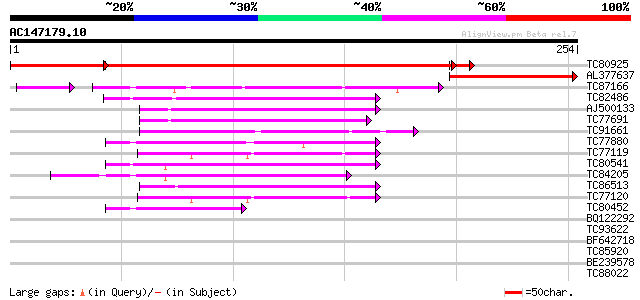
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147179.10 - phase: 1 /pseudo
(254 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC80925 similar to GP|22022575|gb|AAM83244.1 AT3g03330/T21P5_25 ... 310 e-109
AL377637 similar to GP|22022575|gb| AT3g03330/T21P5_25 {Arabidop... 117 6e-27
TC87166 weakly similar to GP|15824408|gb|AAL09328.1 steroleosin ... 67 8e-13
TC82486 weakly similar to SP|P50165|TRNH_DATST Tropinone reducta... 59 2e-09
AJ500133 weakly similar to PIR|T50009|T500 hypothetical protein ... 54 4e-08
TC77691 similar to SP|P28643|FABG_CUPLA 3-oxoacyl-[acyl-carrier ... 52 2e-07
TC91661 similar to GP|14334866|gb|AAK59611.1 unknown protein {Ar... 51 4e-07
TC77880 similar to GP|9622153|gb|AAF89645.1| seed maturation pro... 49 1e-06
TC77119 similar to PIR|D96533|D96533 ARP protein [imported] - Ar... 46 2e-05
TC80541 similar to GP|9758903|dbj|BAB09479.1 Brn1-like protein {... 45 3e-05
TC84205 similar to PIR|T11579|T11579 probable short chain alcoho... 45 3e-05
TC86513 weakly similar to GP|15777867|gb|AAL05894.1 At1g07440/F2... 44 6e-05
TC77120 similar to PIR|D96533|D96533 ARP protein [imported] - Ar... 44 8e-05
TC80452 similar to GP|9622153|gb|AAF89645.1| seed maturation pro... 42 3e-04
BQ122292 similar to GP|9622153|gb| seed maturation protein PM34 ... 36 0.016
TC93622 weakly similar to GP|14023235|dbj|BAB49842. 3-hydroxyacy... 32 0.18
BF642718 similar to SP|P51659|DHB4 Estradiol 17 beta-dehydrogena... 32 0.31
TC85920 homologue to SP|P46256|ALF1_PEA Fructose-bisphosphate al... 30 0.90
BE239578 similar to PIR|T47514|T475 hypothetical protein T6D9.70... 29 1.5
TC88022 weakly similar to PIR|T50009|T50009 hypothetical protein... 29 2.0
>TC80925 similar to GP|22022575|gb|AAM83244.1 AT3g03330/T21P5_25
{Arabidopsis thaliana}, partial (70%)
Length = 854
Score = 310 bits (795), Expect(3) = e-109
Identities = 156/157 (99%), Positives = 157/157 (99%)
Frame = +2
Query: 44 RKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTP 103
RKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTP
Sbjct: 359 RKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTP 538
Query: 104 FMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGP 163
FMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGP
Sbjct: 539 FMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGP 718
Query: 164 IETANNSGSQVPSEKRVSAEKCVELTIIAATHGLKEA 200
IETANNSGSQVPSEKRVSAEKCVELTIIAATHGLKE+
Sbjct: 719 IETANNSGSQVPSEKRVSAEKCVELTIIAATHGLKES 829
Score = 99.4 bits (246), Expect(3) = e-109
Identities = 44/44 (100%), Positives = 44/44 (100%)
Frame = +1
Query: 1 FLDYWCQSWNWGNSSSTTCKFRGQVNTICKG*S*PKPSKVTAKR 44
FLDYWCQSWNWGNSSSTTCKFRGQVNTICKG*S*PKPSKVTAKR
Sbjct: 166 FLDYWCQSWNWGNSSSTTCKFRGQVNTICKG*S*PKPSKVTAKR 297
Score = 23.1 bits (48), Expect(3) = e-109
Identities = 8/11 (72%), Positives = 10/11 (90%)
Frame = +3
Query: 198 KEAWISYQPVL 208
++AWISY PVL
Sbjct: 822 RKAWISYHPVL 854
>AL377637 similar to GP|22022575|gb| AT3g03330/T21P5_25 {Arabidopsis
thaliana}, partial (16%)
Length = 291
Score = 117 bits (292), Expect = 6e-27
Identities = 56/57 (98%), Positives = 57/57 (99%)
Frame = +3
Query: 198 KEAWISYQPVLAVMYLVQYMPTIGYWLMDKVGKNRVEAAKEKGNAYSLSLLFGKKKA 254
KEAWISYQPVLAVMYLVQYMPTIGYWLMDKVGKNRVEAAKEKG+AYSLSLLFGKKKA
Sbjct: 3 KEAWISYQPVLAVMYLVQYMPTIGYWLMDKVGKNRVEAAKEKGHAYSLSLLFGKKKA 173
>TC87166 weakly similar to GP|15824408|gb|AAL09328.1 steroleosin {Sesamum
indicum}, partial (83%)
Length = 1166
Score = 66.6 bits (161), Expect(2) = 8e-13
Identities = 51/160 (31%), Positives = 85/160 (52%), Gaps = 3/160 (1%)
Frame = +1
Query: 38 SKVTAKRKVVDEAESLFPDSGVDYMIHNAAYERPK--SSVLDVTEESLKATFDVNVFGTI 95
SKV R++VDE + F +D++++NAA V D+T + + D N +G++
Sbjct: 325 SKVDDCRRLVDETVNHF--GRLDHLVNNAAISAAMMFEGVTDIT--NWRPLMDTNFWGSV 492
Query: 96 TLTRLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQ 155
TR LR +G VV+SS PAP +++Y+ASK AL + +LR E+ +
Sbjct: 493 YTTRFALTH-LRNSRGKIVVLSSIDSWMPAPRRSIYNASKAALVSLYETLRVEV-GADVG 666
Query: 156 VTVVCPGPIETANNSGS-QVPSEKRVSAEKCVELTIIAAT 194
VT+V PG IE+ G +P E ++ ++ + ++AT
Sbjct: 667 VTIVTPGYIESELTKGKVLLPPEGKMGVDQDMRDVEVSAT 786
Score = 23.5 bits (49), Expect(2) = 8e-13
Identities = 8/26 (30%), Positives = 11/26 (41%)
Frame = +3
Query: 4 YWCQSWNWGNSSSTTCKFRGQVNTIC 29
+WC W W C R ++T C
Sbjct: 162 HWCFLWYWRAFGLRVC*ERCTISTFC 239
>TC82486 weakly similar to SP|P50165|TRNH_DATST Tropinone reductase homolog
(EC 1.1.1.-) (P29X). [Jimsonweed Common thornapple]
{Datura stramonium}, partial (68%)
Length = 653
Score = 58.9 bits (141), Expect = 2e-09
Identities = 37/124 (29%), Positives = 66/124 (52%)
Frame = +3
Query: 43 KRKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLT 102
++K+++ S+F D ++ +++NA PK+ +L+ T E + T +N + L +L
Sbjct: 240 RQKLMETVASIF-DGKLNILVNNAGTITPKT-MLEHTAEDVTNTMGINFESSYHLCQLAH 413
Query: 103 PFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPG 162
P + G G V +SS G P P ++Y+ASK A+N ++ E + I+ VV PG
Sbjct: 414 PLLKESGYGSIVSISSILGLRPLPLCSIYAASKGAINQCTKNIALEYGKDNIRANVVAPG 593
Query: 163 PIET 166
+ T
Sbjct: 594 AVMT 605
>AJ500133 weakly similar to PIR|T50009|T500 hypothetical protein T31P16.40 -
Arabidopsis thaliana, partial (32%)
Length = 623
Score = 54.3 bits (129), Expect = 4e-08
Identities = 30/108 (27%), Positives = 54/108 (49%)
Frame = +3
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +++NA + ++ ++++ T ++N G + + + + P M + G V + S
Sbjct: 261 IDILVNNAGIGST-GPLAELPLDTIRKTLEINTLGQLRMVQQVVPHMALKKSGTIVNVGS 437
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIET 166
G P Y ASK A++ +SLR EL GI V +V PG I +
Sbjct: 438 VVGNISTPWAGSYCASKSAIHAMSNSLRLELKPFGINVVLVMPGSIRS 581
>TC77691 similar to SP|P28643|FABG_CUPLA 3-oxoacyl-[acyl-carrier protein]
reductase chloroplast precursor (EC 1.1.1.100), partial
(82%)
Length = 1481
Score = 52.0 bits (123), Expect = 2e-07
Identities = 30/104 (28%), Positives = 50/104 (47%)
Frame = +2
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +I+NA R ++ + + + D+N+ G T+ M++ KG + +SS
Sbjct: 635 IDVLINNAGITRD-GLLMRMKKSQWQEVIDLNLTGVFLSTQAAAKIMMKNKKGRIINISS 811
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPG 162
G GQA Y+A+K + G S+ E +GI V V PG
Sbjct: 812 VVGLIGNAGQANYAAAKAGVIGLTKSVAKEYSSRGITVNAVAPG 943
>TC91661 similar to GP|14334866|gb|AAK59611.1 unknown protein {Arabidopsis
thaliana}, partial (98%)
Length = 1095
Score = 51.2 bits (121), Expect = 4e-07
Identities = 37/125 (29%), Positives = 59/125 (46%)
Frame = +3
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D ++ NAA S+L + L +++NV TI L + P++ + V++SS
Sbjct: 339 IDVVVSNAAANPSVDSILQTQDSVLDKLWEINVKATILLLKDAAPYLPKGSS--VVIISS 512
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIETANNSGSQVPSEK 178
AG P A+Y +K AL G +L E+ K +V V PG + T N S + S
Sbjct: 513 IAGYHPPASMAMYGVTKTALLGLTKALAGEMAPK-TRVNCVAPGFVPT--NFASFITSNS 683
Query: 179 RVSAE 183
+ E
Sbjct: 684 AMREE 698
>TC77880 similar to GP|9622153|gb|AAF89645.1| seed maturation protein PM34
{Glycine max}, partial (98%)
Length = 1112
Score = 49.3 bits (116), Expect = 1e-06
Identities = 33/124 (26%), Positives = 63/124 (50%), Gaps = 1/124 (0%)
Frame = +1
Query: 44 RKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTP 103
++V+DE + + +D +++NAA + SV ++ E L+ F N+F +TR
Sbjct: 418 KRVIDEIINAY--GRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNIFSYFFMTRHALK 591
Query: 104 FMLRRGKGHFVVMSSAAGKTPAPGQAV-YSASKYALNGYFHSLRSELCQKGIQVTVVCPG 162
M +G ++ +++ + Y+++K A+ + +L +L KGI+V V PG
Sbjct: 592 HMK---EGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLVSKGIRVNGVAPG 762
Query: 163 PIET 166
PI T
Sbjct: 763 PIWT 774
>TC77119 similar to PIR|D96533|D96533 ARP protein [imported] - Arabidopsis
thaliana, partial (93%)
Length = 2277
Score = 45.8 bits (107), Expect = 2e-05
Identities = 40/113 (35%), Positives = 55/113 (48%), Gaps = 4/113 (3%)
Frame = +1
Query: 58 GVDYMIHNAAYERPKSSVLDVTE--ESLKATFDVNVFGTITLTRLLTPFM--LRRGKGHF 113
G+D I +A E P D T+ S + T +VN TRL M L+R G
Sbjct: 367 GLDICIISAGIENPIPFDKDQTDGTRSWRHTLNVNFIAVFDTTRLAIKTMEALKR-PGAI 543
Query: 114 VVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIET 166
+ M SA+G P +YS SK + + SLR +KGI+V V+CP +ET
Sbjct: 544 INMGSASGLYPMYLDPIYSGSKGGVVMFTRSLRLYK-RKGIRVNVLCPEFVET 699
>TC80541 similar to GP|9758903|dbj|BAB09479.1 Brn1-like protein {Arabidopsis
thaliana}, partial (89%)
Length = 1124
Score = 45.1 bits (105), Expect = 3e-05
Identities = 31/124 (25%), Positives = 53/124 (42%), Gaps = 1/124 (0%)
Frame = +3
Query: 44 RKVVDEAESLFPDSGVDYMIHNAAY-ERPKSSVLDVTEESLKATFDVNVFGTITLTRLLT 102
+ + D AE F +S V ++++A + ++ + T E+ +VN G +
Sbjct: 336 KSLFDSAEQAF-NSPVHILVNSAGVLDAELPTIANTTVETFDRIMNVNARGAFLCAKEAA 512
Query: 103 PFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPG 162
+ R G G + ++++ PG Y+ASK + L EL GI V PG
Sbjct: 513 NRLKRGGGGRIIFLTTSLAAAFKPGYGAYTASKAGVEAMTKILAKELKGTGITANCVAPG 692
Query: 163 PIET 166
P T
Sbjct: 693 PTAT 704
>TC84205 similar to PIR|T11579|T11579 probable short chain alcohol
dehydrogenase CPRD12 drought-inducible - cowpea,
partial (61%)
Length = 645
Score = 45.1 bits (105), Expect = 3e-05
Identities = 40/137 (29%), Positives = 60/137 (43%), Gaps = 2/137 (1%)
Frame = +1
Query: 19 CKFRGQVNTICKG*S*PKPSKVTAKRKVVDEAESLFPDSGVDYMIHNAAY--ERPKSSVL 76
CK G N +C S + + VVD A S + +D M +NA + +L
Sbjct: 145 CKNLGTKNILCVHCDVTIESDI---KTVVDIAVSNY--GKLDIMFNNAGI*SDDKNREIL 309
Query: 77 DVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKY 136
+ E+ K FDVNV+G + M+ + KG + +S A K Y+ SK+
Sbjct: 310 NYDSEAFKRVFDVNVYGAFLGAKHAARVMIPQKKGVILFTASVATKIAGETTHAYTCSKH 489
Query: 137 ALNGYFHSLRSELCQKG 153
AL G +L EL + G
Sbjct: 490 ALVGLTKNLCVELGKYG 540
>TC86513 weakly similar to GP|15777867|gb|AAL05894.1 At1g07440/F22G5_16
{Arabidopsis thaliana}, partial (89%)
Length = 1484
Score = 43.9 bits (102), Expect = 6e-05
Identities = 28/108 (25%), Positives = 47/108 (42%)
Frame = +1
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
++ +++N K + LD TE+ + N+ +++L P + V MSS
Sbjct: 298 LNILVNNVGTNMQKQT-LDFTEQDFSFLVNTNLESAFHISQLAHPLLKASNNASIVFMSS 474
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIET 166
G +YSA+K A+ +L E + I+ V PGPI T
Sbjct: 475 IGGVASLNIGTIYSAAKGAIIQLTKNLACEWAKDNIRTNCVAPGPIRT 618
>TC77120 similar to PIR|D96533|D96533 ARP protein [imported] - Arabidopsis
thaliana, partial (93%)
Length = 2203
Score = 43.5 bits (101), Expect = 8e-05
Identities = 38/113 (33%), Positives = 54/113 (47%), Gaps = 4/113 (3%)
Frame = +3
Query: 58 GVDYMIHNAAYERPKSSVLDVTE--ESLKATFDVNVFGTITLTRLLTPFM--LRRGKGHF 113
G+D I +A P D T+ S + T +VN TRL M L+R G
Sbjct: 393 GLDICIASAGINNPIPFDKDPTDGTRSWRHTLNVNFIAVFDTTRLAIKAMEALKR-PGTI 569
Query: 114 VVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIET 166
+ + SA+G P G VY+ SK + + +LR Q GI++ V+CP IET
Sbjct: 570 INLGSASGLYPMYGDPVYTGSKGGVVMFTRALRLYKRQ-GIRINVLCPEFIET 725
>TC80452 similar to GP|9622153|gb|AAF89645.1| seed maturation protein PM34
{Glycine max}, complete
Length = 1257
Score = 41.6 bits (96), Expect = 3e-04
Identities = 22/63 (34%), Positives = 37/63 (57%)
Frame = +3
Query: 44 RKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTP 103
+KVVDE + + +D +++NAA + SSV ++ E L+ F N+F +TR +TP
Sbjct: 411 KKVVDEIVNAY--GHIDILVNNAAEQYECSSVEEIDESRLERVFRTNIFSYFFMTRSVTP 584
Query: 104 FML 106
+L
Sbjct: 585 TLL 593
Score = 31.2 bits (69), Expect = 0.40
Identities = 15/36 (41%), Positives = 22/36 (60%)
Frame = +1
Query: 131 YSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIET 166
Y+++K A+ + L +L KGI+V V PGPI T
Sbjct: 802 YTSTKGAIVAFTRGLSLQLVSKGIRVNGVAPGPIWT 909
>BQ122292 similar to GP|9622153|gb| seed maturation protein PM34 {Glycine
max}, partial (48%)
Length = 437
Score = 35.8 bits (81), Expect = 0.016
Identities = 14/43 (32%), Positives = 27/43 (62%)
Frame = +2
Query: 57 SGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTR 99
S +D +++NAA + ++S+ ++TE+ L+ F N+F L R
Sbjct: 263 SSIDVLVNNAAEQHLRNSIEEITEQQLERVFRTNIFSHFFLVR 391
>TC93622 weakly similar to GP|14023235|dbj|BAB49842. 3-hydroxyacyl-CoA
dehydrogenase type II {Mesorhizobium loti}, partial
(30%)
Length = 460
Score = 32.3 bits (72), Expect = 0.18
Identities = 19/57 (33%), Positives = 29/57 (50%)
Frame = +3
Query: 110 KGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIET 166
+G + +SS A + GQ YSA+K + + +L + GI+V V PG ET
Sbjct: 54 RGVIINISSIAYQDGQVGQVTYSATKGGVASMTLPMTRDLSRFGIRVVAVAPGLFET 224
>BF642718 similar to SP|P51659|DHB4 Estradiol 17 beta-dehydrogenase 4 (EC
1.1.1.62) (17-beta-HSD 4), partial (24%)
Length = 569
Score = 31.6 bits (70), Expect = 0.31
Identities = 25/90 (27%), Positives = 42/90 (45%)
Frame = +1
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +I+NA R +S ++++ V++ G T ++ G ++ SS
Sbjct: 274 IDILINNAGILRDRSFT-KMSDQDWDLIHLVHLKGAFKTTHAAWEHFRKQKYGRVIMTSS 450
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSE 148
AG GQA YSA+K L G ++L E
Sbjct: 451 NAGIFGNFGQANYSAAKMGLVGLTNTLAIE 540
>TC85920 homologue to SP|P46256|ALF1_PEA Fructose-bisphosphate aldolase
cytoplasmic isozyme 1 (EC 4.1.2.13). [Garden pea] {Pisum
sativum}, complete
Length = 1253
Score = 30.0 bits (66), Expect = 0.90
Identities = 23/74 (31%), Positives = 35/74 (47%)
Frame = -2
Query: 69 ERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSSAAGKTPAPGQ 128
ER S++ +V S +A ++F T+ L R L V + S+A K P PG
Sbjct: 247 ERYWSALGEVKRSSRRA*RLASMFSTLMLARRLP----------IVPVLSSAAKIPLPGV 98
Query: 129 AVYSASKYALNGYF 142
A+Y A + + YF
Sbjct: 97 AIYLAFLISSSAYF 56
>BE239578 similar to PIR|T47514|T475 hypothetical protein T6D9.70 -
Arabidopsis thaliana, partial (26%)
Length = 600
Score = 29.3 bits (64), Expect = 1.5
Identities = 14/67 (20%), Positives = 34/67 (49%)
Frame = -2
Query: 126 PGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIETANNSGSQVPSEKRVSAEKC 185
P Q +Y ++G+F+S+ +++C K + + + P+E ++ ++ P +S
Sbjct: 371 PNQLSTFLCQYVVHGFFYSVNNQICWKFVMIFL----PLENTSSCENRKPRMPGLSCTSD 204
Query: 186 VELTIIA 192
V I++
Sbjct: 203 VNCRIVS 183
>TC88022 weakly similar to PIR|T50009|T50009 hypothetical protein T31P16.40
- Arabidopsis thaliana, partial (87%)
Length = 1093
Score = 28.9 bits (63), Expect = 2.0
Identities = 13/53 (24%), Positives = 29/53 (54%)
Frame = +3
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKG 111
+D +++NA + ++ +++ TF+ NVFG++ + + + P M R G
Sbjct: 264 IDVLVNNAGVPCV-GPLAEIPLSAIQNTFETNVFGSMRMVQAVVPHMATRKPG 419
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.134 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,062,127
Number of Sequences: 36976
Number of extensions: 102016
Number of successful extensions: 575
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 569
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 572
length of query: 254
length of database: 9,014,727
effective HSP length: 94
effective length of query: 160
effective length of database: 5,538,983
effective search space: 886237280
effective search space used: 886237280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC147179.10


