

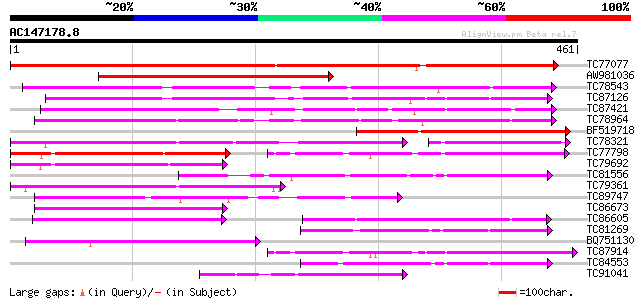
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147178.8 + phase: 0
(461 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77077 similar to GP|9293909|dbj|BAB01812.1 sugar transporter p... 442 e-124
AW981036 weakly similar to PIR|G84564|G84 probable sugar transpo... 377 e-105
TC78543 similar to PIR|T14545|T14545 probable sugar transporter ... 184 6e-47
TC87126 similar to GP|15724240|gb|AAL06513.1 At1g75220/F22H5_6 {... 175 4e-44
TC87421 sugar transporter 162 3e-40
TC78964 weakly similar to PIR|T00450|T00450 probable monosacchar... 154 5e-38
BF519718 weakly similar to PIR|G84564|G84 probable sugar transpo... 154 9e-38
TC78321 similar to GP|21689841|gb|AAM67564.1 unknown protein {Ar... 141 5e-34
TC77798 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_14... 119 2e-27
TC79692 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_14... 113 2e-25
TC81556 similar to GP|2688830|gb|AAB88879.1| putative sugar tran... 100 1e-21
TC79361 similar to GP|17380890|gb|AAL36257.1 putative membrane t... 100 2e-21
TC89747 similar to PIR|F96696|F96696 protein F1N21.12 [imported]... 99 4e-21
TC86673 weakly similar to PIR|S25015|S25015 monosaccharide trans... 98 8e-21
TC86605 similar to PIR|T01853|T01853 probable hexose transport p... 90 2e-18
TC81269 similar to GP|23504385|emb|CAC00697. putative sugar tran... 89 5e-18
BQ751130 weakly similar to GP|15289796|dbj putative monosacchari... 88 6e-18
TC87914 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_14... 87 1e-17
TC84553 similar to GP|17381265|gb|AAL36051.1 AT5g18840/F17K4_90 ... 84 9e-17
TC91041 similar to GP|21689841|gb|AAM67564.1 unknown protein {Ar... 78 8e-15
>TC77077 similar to GP|9293909|dbj|BAB01812.1 sugar transporter protein
{Arabidopsis thaliana}, partial (85%)
Length = 1752
Score = 442 bits (1138), Expect = e-124
Identities = 224/448 (50%), Positives = 314/448 (70%), Gaps = 2/448 (0%)
Frame = +1
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA ++IK +L++SD ++++L GI+N +L +AGR SD+IGRRYTI+ + FF+G
Sbjct: 172 MSGAAIYIKRDLKVSDGKIEVLLGIINIYSLIGSCLAGRTSDWIGRRYTIVFAGAIFFVG 351
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
++LMG+ ++ LM GR +AG G+G+AL+I VY+AE+S S RGFLTS P++ IN G L
Sbjct: 352 ALLMGFSPNYNFLMFGRFVAGVGIGYALMIAPVYTAEVSPASSRGFLTSFPEVFINSGIL 531
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SNY KLSL+LGWR+ML + +IPS+ L + +L + ESPRWLVM+GRLGDA KVL
Sbjct: 532 LGYISNYAFSKLSLKLGWRMMLGVGAIPSVILAVGVLAMPESPRWLVMRGRLGDAIKVLN 711
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
S+SK+EA+ R+ EIK A GI E+C ++V V K +G G KE+F P+P V I+I
Sbjct: 712 KTSDSKEEAQLRLAEIKQAAGIPEDCNDDVVEVKVK-NTGEGVWKELFLYPTPAVRHIVI 888
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
AA+G+H FQ GV+ + LYSP IF + GI LL+AT+ +G +TLF L++ F+LD+
Sbjct: 889 AALGIHFFQQASGVDAVVLYSPTIFKKAGINGDTHLLIATIAVGFVKTLFILVATFMLDR 1068
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVE--NSKLGEEPLWAIIFTIIVIYIMAGFNAIG 358
GRR LLL S GG++ S+L L V I++ N+KL WAI +I + +IG
Sbjct: 1069YGRRPLLLTSVGGMVVSLLTLAVSLTIIDHSNTKLN----WAIGLSIATVLSYVATFSIG 1236
Query: 359 IGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTN 418
G +TWVYS+EIFPLRLRAQG V++NR+T+ + +F+S+ K IT+GG FFLF G
Sbjct: 1237AGPITWVYSSEIFPLRLRAQGAACGVVVNRVTSGVISMTFLSLSKGITIGGAFFLFGGIA 1416
Query: 419 VLGWWFYYSFLPETKGRSLEDMETIFGK 446
+GW F+Y LPET+G++LE+ME FGK
Sbjct: 1417TIGWIFFYIMLPETQGKTLEEMEASFGK 1500
>AW981036 weakly similar to PIR|G84564|G84 probable sugar transporter
[imported] - Arabidopsis thaliana, partial (25%)
Length = 575
Score = 377 bits (967), Expect = e-105
Identities = 191/191 (100%), Positives = 191/191 (100%)
Frame = +2
Query: 73 LMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGKL 132
LMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGKL
Sbjct: 2 LMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGKL 181
Query: 133 SLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQR 192
SLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQR
Sbjct: 182 SLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQR 361
Query: 193 MKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNIC 252
MKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNIC
Sbjct: 362 MKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNIC 541
Query: 253 GVEGIFLYSPR 263
GVEGIFLYSPR
Sbjct: 542 GVEGIFLYSPR 574
>TC78543 similar to PIR|T14545|T14545 probable sugar transporter protein -
beet, partial (93%)
Length = 1653
Score = 184 bits (467), Expect = 6e-47
Identities = 121/436 (27%), Positives = 219/436 (49%), Gaps = 2/436 (0%)
Frame = +1
Query: 11 ELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYGSSF 70
+L +S + L + N A+ + +G++++Y+GR+ ++M++SI +G +++ + +
Sbjct: 241 DLGLSVSEFSLFGSLSNVGAMVGAIASGQIAEYMGRKGSLMIASIPNIIGWLMISFANDS 420
Query: 71 PILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLG 130
L +GR + GFGVG V VY AEIS + RG L S+ LS+ +G +L YL
Sbjct: 421 SFLYMGRLLEGFGVGIISYTVPVYIAEISPQNLRGSLVSVNQLSVTLGIMLAYL------ 582
Query: 131 KLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAE 190
L L + WR + + IP L+ + + ESPRWL G + + L ++ + +
Sbjct: 583 -LGLFVEWRFLAILGIIPCTLLIPGLFFIPESPRWLAKMGMTEEFENSLQVLRGFETDIS 759
Query: 191 QRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQN 250
+ EIK AV +++T LK+ Y + L+ IG+ + Q
Sbjct: 760 VEVNEIKTAV----------ASANRRTTVRFSELKQRRY------WLPLMIGIGLLVLQQ 891
Query: 251 ICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRILLLVS 310
+ G+ G+ YS IF GI+ +AT G+G Q L T L+ +L DK GRR+LL+VS
Sbjct: 892 LSGINGVLFYSSTIFQNAGISSSD---VATFGVGAVQVLATTLTLWLADKSGRRLLLIVS 1062
Query: 311 SGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIV--IYIMAGFNAIGIGAVTWVYST 368
S + S+L + + + + + L+ I+ + V + +M ++G+GA+ W+ +
Sbjct: 1063SSAMTLSLLVVSISFYLKDYYISADSSLYGILSLLSVAGVVVMVIAFSLGMGAMPWIIMS 1242
Query: 369 EIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYYSF 428
EI P+ ++ + N + ++T ++ + GGTF ++ F +
Sbjct: 1243EILPINIKGLAGSFATLANWFFS-WLITLTANLLLDWSSGGTFTIYTVVCAFTVGFVAIW 1419
Query: 429 LPETKGRSLEDMETIF 444
+PETKG++LE+++ F
Sbjct: 1420VPETKGKTLEEIQQFF 1467
>TC87126 similar to GP|15724240|gb|AAL06513.1 At1g75220/F22H5_6 {Arabidopsis
thaliana}, partial (79%)
Length = 1524
Score = 175 bits (443), Expect = 4e-44
Identities = 116/416 (27%), Positives = 210/416 (49%), Gaps = 4/416 (0%)
Frame = +3
Query: 30 ALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYGSSFPILMIGRCIAGFGVGFALI 89
A+ + +G++++Y+GR+ ++M++SI +G + + + L +GR + GFGVG
Sbjct: 9 AMVGAIASGQIAEYVGRKGSLMIASIPNIIGWLAISFAKDSSFLFMGRLLEGFGVGIISY 188
Query: 90 IVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGKLSLRLGWRIMLAIPSIPS 149
+V VY AEI+ + RG L S+ LS+ IG +L YL L L WR++ + +P
Sbjct: 189 VVPVYIAEIAPENMRGSLGSVNQLSVTIGIMLAYL-------LGLFANWRVLAILGILPC 347
Query: 150 IGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQRMKEIKNAVGID-ENCTQ 208
L+ + + ESPRWL G + + + L ++ + + EIK AV + + T
Sbjct: 348 TVLIPGLFFIPESPRWLAKMGMMEEFETSLQVLRGFDTDISVEVHEIKKAVASNGKRATI 527
Query: 209 NIVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNICGVEGIFLYSPRIFGRM 268
+ +K ++ P L IG+ + Q + G+ G+ YS IF
Sbjct: 528 RFADLQRK----------RYWFP-------LSVGIGLLVLQQLSGINGVLFYSTSIFANA 656
Query: 269 GITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVIFSMLGLCVC---S 325
GI+ ATVG+G Q + T ++ +L+DK GRR+LL++SS + S+L + +
Sbjct: 657 GISSSNA---ATVGLGAIQVIATGVATWLVDKSGRRVLLIISSSLMTASLLVVSIAFYLE 827
Query: 326 AIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPLRLRAQGLGVCVI 385
+VE L II + ++ ++ GF ++G+G + W+ +EI P+ ++ +
Sbjct: 828 GVVEKDSQYFSIL-GIISVVGLVVMVIGF-SLGLGPIPWLIMSEILPVNIKGLAGSTATM 1001
Query: 386 MNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYYSFLPETKGRSLEDME 441
N + ++T ++ T + GGTF ++ F ++PETKGR+LE+++
Sbjct: 1002ANWLV-AWIITMTANLLLTWSSGGTFLIYTVVAAFTVVFTSLWVPETKGRTLEEIQ 1166
>TC87421 sugar transporter
Length = 1753
Score = 162 bits (410), Expect = 3e-40
Identities = 120/427 (28%), Positives = 206/427 (48%), Gaps = 8/427 (1%)
Frame = +2
Query: 26 LNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYGSSFPILMIGRCIAGFGVG 85
L AL + ++A ++ GR+ +++ + F +G+++ G+ + +L++GR + GFG+G
Sbjct: 293 LYLAALLSSLVASTITRRFGRKLSMLFGGLLFLVGALINGFANHVWMLIVGRILLGFGIG 472
Query: 86 FALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGKLSLRLGWRIMLAIP 145
FA V +Y +E++ YRG L LSI IG L+ + NYF K+ GWR+ L
Sbjct: 473 FANQPVPLYLSEMAPYKYRGALNIGFQLSITIGILVANVLNYFFAKIKGGWGWRLSLGGA 652
Query: 146 SIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQRMKEIKNAVGIDEN 205
+P++ + I L L ++P ++ +G + A+ ++K I+ +DE
Sbjct: 653 MVPALIITIGSLVLPDTPNSMIERG--------------DRDGAKAQLKRIRGIEDVDEE 790
Query: 206 CTQNIV--HVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNICGVEGIFLYSPR 263
+ S + + L + Y+P L A+ + FQ G+ I Y+P
Sbjct: 791 FNDLVAASEASMQVENPWRNLLQRKYRPQ------LTMAVLIPFFQQFTGINVIMFYAPV 952
Query: 264 IFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVIFSMLGLCV 323
+F +G D +L+ A + G+ + T +S + +DK GRR L L GG + + V
Sbjct: 953 LFNSIGFKDDASLMSAVI-TGVVNVVATCVSIYGVDKWGRRALFL--EGGAQMLICQVAV 1123
Query: 324 CSAI----VENSKLGEEPLW-AIIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPLRLRAQ 378
+AI + G P W AI+ + + +AGF A G + W+ +EIFPL +R+
Sbjct: 1124AAAIGAKFGTSGNPGNLPEWYAIVVVLFICIYVAGF-AWSWGPLGWLVPSEIFPLEIRSA 1300
Query: 379 GLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYYSF-LPETKGRSL 437
V V +N + V F+ + + G FLF VL Y F LPETKG +
Sbjct: 1301AQSVNVSVNMLFTFLVAQVFLIMLCHMKFG--LFLFFAFFVLVMSIYVFFLLPETKGIPI 1474
Query: 438 EDMETIF 444
E+M+ ++
Sbjct: 1475EEMDRVW 1495
>TC78964 weakly similar to PIR|T00450|T00450 probable monosaccharide
transport protein T14N5.7 - Arabidopsis thaliana,
partial (98%)
Length = 1771
Score = 154 bits (390), Expect = 5e-38
Identities = 121/427 (28%), Positives = 201/427 (46%), Gaps = 3/427 (0%)
Frame = +1
Query: 21 LLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYGSSFPILMIGRCIA 80
L L AL A L+ GR+ TI++ ++ F +G+IL + P L+IGR
Sbjct: 289 LFTSSLYFSALVMTFFASYLTRNKGRKATIIVGALSFLIGAILNAAAQNIPTLIIGRVFL 468
Query: 81 GFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGKLSLRLGWRI 140
G G+GF V +Y +E++ S RG + L + G L+ L NYF K+ GWRI
Sbjct: 469 GGGIGFGNQAVPLYLSEMAPASSRGAVNQLFQFTTCAGILIANLVNYFTDKIHPH-GWRI 645
Query: 141 MLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQRMKEIKNAV 200
L + IP++ +++ + E+P LV QGRL +A+KVL + +K + +++K+A
Sbjct: 646 SLGLAGIPAVLMLLGGIFCAETPNSLVEQGRLDEARKVLEKVRGTK-NVDAEFEDLKDA- 819
Query: 201 GIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNICGVEGIFLY 260
+++ +S L + Y+P +++I A+G FQ + G I Y
Sbjct: 820 ----------SELAQAVKSPFKVLLKRKYRP-----QLIIGALGTPAFQQLTGNNSILFY 954
Query: 261 SPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVIFSMLG 320
+P IF G L + + G + + T++S FL+DK G R L + F M+
Sbjct: 955 APVIFQSSGFGSNAALFSSFITNG-ALLVATVISMFLVDKFGTRKFFLEAG----FEMIC 1119
Query: 321 LCVCSAIVENSKLG---EEPLWAIIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPLRLRA 377
+ +A+V + G E F +I+I+ G + W+ +E+FPL +R+
Sbjct: 1120CMIITAVVLAVEFGHGKELSKGISAFLVIMIFWFVLAYGRSWGPLGWLVPSELFPLEIRS 1299
Query: 378 QGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYYSFLPETKGRSL 437
+ V +N I V F+ + G F LF G V+ F + LPETK +
Sbjct: 1300AAQSIAVCVNMIFTALVAQLFLLSLCHLKY-GIFLLFGGLIVVMSVFVFFLLPETKQVPI 1476
Query: 438 EDMETIF 444
E++ +F
Sbjct: 1477EEIYLLF 1497
>BF519718 weakly similar to PIR|G84564|G84 probable sugar transporter
[imported] - Arabidopsis thaliana, partial (20%)
Length = 659
Score = 154 bits (388), Expect = 9e-38
Identities = 75/174 (43%), Positives = 114/174 (65%)
Frame = +3
Query: 283 IGISQTLFTLLSCFLLDKIGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAII 342
+GI++T F L S +LD+ GRR +LL+ S G+ S+ GL + ++ NS E+P+WAI
Sbjct: 9 MGIAKTCFVLFSALVLDRFGRRPMLLLGSSGMAVSLFGLGMGCTLLHNSD--EKPMWAIA 182
Query: 343 FTIIVIYIMAGFNAIGIGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIY 402
++ + F +IG+G TWVYS+EIFP+RLRAQG + + +NR+ + V SF+SI
Sbjct: 183 LCVVAVCAAVSFFSIGLGPTTWVYSSEIFPMRLRAQGTSLAISVNRLISGVVSMSFLSIS 362
Query: 403 KTITLGGTFFLFVGTNVLGWWFYYSFLPETKGRSLEDMETIFGKNSNSEVQMKY 456
+ IT GG FF+ G VL F+Y FLPETKG+SLE++E +F + + ++ K+
Sbjct: 363 EEITFGGMFFVLAGVMVLATLFFYYFLPETKGKSLEEIEALFEEELS*NLETKF 524
>TC78321 similar to GP|21689841|gb|AAM67564.1 unknown protein {Arabidopsis
thaliana}, partial (80%)
Length = 2093
Score = 141 bits (356), Expect = 5e-34
Identities = 102/327 (31%), Positives = 162/327 (49%), Gaps = 4/327 (1%)
Frame = +2
Query: 1 MAGALLFIKEELEISDMQVQLLAGILN---ACALPACMIAGRLSDYIGRRYTIMLSSIFF 57
++GALL+I++E + + L I++ A A+ I G ++D GR+ +I+++ F
Sbjct: 284 ISGALLYIRDEFPAVEKKTWLQEAIVSTAIAGAIIGAAIGGWINDRFGRKVSIIVADTLF 463
Query: 58 FLGSILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINI 117
LGSI++ + L++GR G GVG A + +Y +E S RG L SL I
Sbjct: 464 LLGSIILAAAPNPATLIVGRVFVGLGVGMASMASPLYISEASPTRVRGALVSLNSFLITG 643
Query: 118 GFLLGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKK 177
G L YL N K WR ML + + P++ ++LML L ESPRWL +G+ +AK
Sbjct: 644 GQFLSYLINLAFTKAPGT--WRWMLGVAAAPAVIQIVLMLSLPESPRWLYRKGKEEEAKV 817
Query: 178 VLLLISNSKQEAEQRMKEIKNAVGIDENCTQNI-VHVSKKTRSGGGALKEMFYKPSPHVY 236
+L I ++ + ++ +K +V ++ T+ I + KT S V
Sbjct: 818 ILKKI-YEVEDYDNEIQALKESVEMELKETEKISIMQLVKTTS---------------VR 949
Query: 237 RILIAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCF 296
R L A +G+ FQ G+ + YSP I G K T LL ++ ++LS +
Sbjct: 950 RGLYAGVGLAFFQQFTGINTVMYYSPSIVQLAGFASKRTALLLSLITSGLNAFGSILSIY 1129
Query: 297 LLDKIGRRILLLVSSGGVIFSMLGLCV 323
+DK GR+ L L+S GV+ S+ L V
Sbjct: 1130FIDKTGRKKLALISLTGVVLSLTLLTV 1210
Score = 62.4 bits (150), Expect = 4e-10
Identities = 38/116 (32%), Positives = 64/116 (54%)
Frame = +2
Query: 341 IIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFIS 400
I + +YI+ F + G+G V WV ++EI+PLR R G+ ++N+ V SF+S
Sbjct: 1490 IAILALALYII--FFSPGMGTVPWVVNSEIYPLRYRGICGGIASTTVWVSNLVVSQSFLS 1663
Query: 401 IYKTITLGGTFFLFVGTNVLGWWFYYSFLPETKGRSLEDMETIFGKNSNSEVQMKY 456
+ I TF +F ++ +F F+PETKG +E++E++ K VQ+K+
Sbjct: 1664 LTVAIGPAWTFMIFAIIAIVAIFFVIIFVPETKGVPMEEVESMLEKRF---VQIKF 1822
>TC77798 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_140
{Arabidopsis thaliana}, partial (88%)
Length = 2467
Score = 119 bits (299), Expect = 2e-27
Identities = 69/182 (37%), Positives = 112/182 (60%), Gaps = 3/182 (1%)
Frame = +3
Query: 1 MAGALLFIKEELEISDMQVQLLAG--ILNACALPACMIAGRLSDYIGRRYTIMLSSIFFF 58
+AG++L+IK++L + L+ ++ A + C +G +SD++GRR +++SS+ +F
Sbjct: 183 IAGSILYIKKDLALQTTMEGLVVAMSLIGATVITTC--SGPISDWLGRRPMMIISSVLYF 356
Query: 59 LGSILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIG 118
LGS++M + + +L + R + GFG+G A+ +V VY +E + RG L +LP S + G
Sbjct: 357 LGSLVMLWSPNVYVLCLARLLDGFGIGLAVTLVPVYISETAPSDIRGSLNTLPQFSGSGG 536
Query: 119 FLLGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVIL-MLQLVESPRWLVMQGRLGDAKK 177
L Y F+ LS WRIML + SIPS+ +L + L ESPRWLV +G++ +AKK
Sbjct: 537 MFLSYCM-VFVMSLSPSPSWRIMLGVLSIPSLFYFLLTVFFLPESPRWLVSKGKMLEAKK 713
Query: 178 VL 179
VL
Sbjct: 714 VL 719
Score = 92.4 bits (228), Expect = 3e-19
Identities = 73/259 (28%), Positives = 118/259 (45%), Gaps = 13/259 (5%)
Frame = +3
Query: 210 IVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNICGVEGIFLYSPRIFGRMG 269
++H SK T S G + + P V LI IG+ + Q G+ G+ Y+P+I G
Sbjct: 1560 MIHPSK-TASKGPIWEALL---EPGVKHALIVGIGIQLLQQFSGINGVLYYTPQILEEAG 1727
Query: 270 ITDKGTLLLATVGIGISQTLFTL-------------LSCFLLDKIGRRILLLVSSGGVIF 316
+ +LLA +G+ + + F + L+ L+D GRR LLLV+ +I
Sbjct: 1728 VA----VLLADLGLSSTSSSFLISAVTTLLMLPSIGLAMRLMDVTGRRQLLLVTIPVLIV 1895
Query: 317 SMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPLRLR 376
S++ L + S I G AI +V+Y F +G G + + +EIFP R+R
Sbjct: 1896 SLVILVLGSVI----DFGSVVHAAISTVCVVVYFC--FFVMGYGPIPNILCSEIFPTRVR 2057
Query: 377 AQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYYSFLPETKGRS 436
+ +C ++ I ++ V S + ++ L G F ++ + W F Y +PETKG
Sbjct: 2058 GLCIAICALVFWIGDIIVTYSLPVMLSSLGLAGVFGVYAIVCCISWVFVYLKVPETKGMP 2237
Query: 437 LEDMETIFGKNSNSEVQMK 455
LE + F S K
Sbjct: 2238 LEVITEFFSVGSKQSAAAK 2294
>TC79692 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_140
{Arabidopsis thaliana}, partial (49%)
Length = 1353
Score = 113 bits (282), Expect = 2e-25
Identities = 67/182 (36%), Positives = 109/182 (59%), Gaps = 5/182 (2%)
Frame = +2
Query: 1 MAGALLFIKEELEI-SDMQVQLLA---GILNACALPACMIAGRLSDYIGRRYTIMLSSIF 56
+AG++L+IK E ++ S+ V+ L ++ A + C +G LSD GRR +++SS+
Sbjct: 224 IAGSILYIKREFQLQSEPTVEGLIVAMSLIGATVVTTC--SGALSDLFGRRPMLIISSLL 397
Query: 57 FFLGSILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSIN 116
+FL S++M + + IL+ R + G G+G A+ +V +Y +EI+ P RG L +LP + +
Sbjct: 398 YFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIAPPEIRGSLNTLPQFAGS 577
Query: 117 IGFLLGYLSNYFLGKLSLRLGWRIMLAIPSIPS-IGLVILMLQLVESPRWLVMQGRLGDA 175
G Y + + L+ WR+ML + SIPS I + +L L ESPRWLV +GR+ +A
Sbjct: 578 AGMFFSYCMVFGM-SLTKAPSWRLMLGVLSIPSLIYFALTLLVLPESPRWLVSKGRMLEA 754
Query: 176 KK 177
KK
Sbjct: 755 KK 760
>TC81556 similar to GP|2688830|gb|AAB88879.1| putative sugar transporter
{Prunus armeniaca}, partial (59%)
Length = 1074
Score = 100 bits (249), Expect = 1e-21
Identities = 84/306 (27%), Positives = 140/306 (45%), Gaps = 2/306 (0%)
Frame = +1
Query: 138 WRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQRMKEIK 197
WR M I +PSI L + M ESPRWL QG++ +A+K IK
Sbjct: 19 WRTMFGIAVVPSILLALGMAISPESPRWLFQQGKIAEAEKA-----------------IK 147
Query: 198 NAVGIDENCTQNIVHVSKKTRSGGGALKEM--FYKPSPHVYRILIAAIGVHIFQNICGVE 255
G + T V +T S G + E F S ++++ + +FQ + G+
Sbjct: 148 TLYGKERVAT---VMYDLRTASQGSSEPEAGWFDLFSSRYWKVVSVGAALFLFQQLAGIN 318
Query: 256 GIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVI 315
+ YS +F GI + A+ +G S + T ++ L+DK GR+ LL+ S G+
Sbjct: 319 AVVYYSTSVFRSAGIASD---VAASALVGASNVIGTAIASSLMDKQGRKSLLITSFSGMA 489
Query: 316 FSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPLRL 375
SML L + + A++ T V+Y+++ ++G G V + EIF R+
Sbjct: 490 ASMLLLSL--SFTWKVLAPYSGTLAVLGT--VLYVLSF--SLGAGPVPALLLPEIFASRI 651
Query: 376 RAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYYSFLPETKGR 435
RA+ + + + + I+N + F+S+ + + F VL + + ETKGR
Sbjct: 652 RAKAVSLSLGTHWISNFVIGLYFLSVVNKFGISSVYLGFSAVCVLAVLYIAGNVVETKGR 831
Query: 436 SLEDME 441
SLE++E
Sbjct: 832 SLEEIE 849
>TC79361 similar to GP|17380890|gb|AAL36257.1 putative membrane transporter
protein {Arabidopsis thaliana}, partial (55%)
Length = 949
Score = 100 bits (248), Expect = 2e-21
Identities = 76/234 (32%), Positives = 115/234 (48%), Gaps = 10/234 (4%)
Frame = +1
Query: 1 MAGALLFIKEE---LEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFF 57
++GALL+IK++ + S+ + + + A A+ G L+D GR+ +L+ + F
Sbjct: 232 ISGALLYIKDDFPQVRNSNFLQETIVSMAIAGAIVGAAFGGWLNDAYGRKKATLLADVIF 411
Query: 58 FLGSILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINI 117
LG+ILM +L+ GR + G GVG A + VY AE++ RG L S L I
Sbjct: 412 ILGAILMAAAPDPYVLIAGRLLVGLGVGIASVTAPVYIAEVAPSEIRGSLVSTNVLMITG 591
Query: 118 GFLLGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKK 177
G + YL N ++ WR ML + +P++ I ML L ESPRWL ++ R +A
Sbjct: 592 GQFVSYLVNLVFTQVPGT--WRWMLGVSGVPALIQFICMLFLPESPRWLFIKNRKNEAVD 765
Query: 178 VLLLISN-SKQEAEQRMKEIKNAVGIDENCTQNIVHV--SKKTR----SGGGAL 224
V+ I + S+ E E ++ T HV SK+TR GGG L
Sbjct: 766 VISKIYDLSRLEDEIDFLTAQSEQERQRRSTIKFWHVFRSKETRLAFLDGGGLL 927
>TC89747 similar to PIR|F96696|F96696 protein F1N21.12 [imported] -
Arabidopsis thaliana, partial (64%)
Length = 1372
Score = 98.6 bits (244), Expect = 4e-21
Identities = 76/304 (25%), Positives = 136/304 (44%), Gaps = 5/304 (1%)
Frame = +3
Query: 21 LLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYGSSFPILMIGRCIA 80
L+ I AL C+++G ++D +GRR L ++ +G+ + ++ +++GR
Sbjct: 501 LVVSICLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAMSAATNNLFGMLVGRLFV 680
Query: 81 GFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGKLSLRLG--W 138
G G+G + ++Y E+S RG +L I I G L + F+G + W
Sbjct: 681 GTGLGLGPPVAALYVTEVSPAFVRGTYGAL----IQIATCFGILGSLFIGIPVKEISGWW 848
Query: 139 RIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAK---KVLLLISNSKQEAEQRMKE 195
R+ + +IP+ L + M ESP WL QGR +A+ + LL +S +K Q K
Sbjct: 849 RVCFWVSTIPAAILALAMDFCAESPHWLYKQGRTAEAEAEFERLLGVSEAKFAMSQLSK- 1025
Query: 196 IKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNICGVE 255
+D + V S+ H +++ + Q + G+
Sbjct: 1026------VDRGEDTDTVKFSELLHG--------------HHSKVVFIGSTLFALQQLSGIN 1145
Query: 256 GIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVI 315
+F +S +F G+ A V IG++ +++S L+DK+GR++LL S G+
Sbjct: 1146AVFYFSSTVFKSAGVPSD----FANVCIGVANLTGSIISTGLMDKLGRKVLLFWSFFGMA 1313
Query: 316 FSML 319
SM+
Sbjct: 1314ISMI 1325
>TC86673 weakly similar to PIR|S25015|S25015 monosaccharide transport
protein MST1 - common tobacco, partial (65%)
Length = 1162
Score = 97.8 bits (242), Expect = 8e-21
Identities = 55/157 (35%), Positives = 87/157 (55%)
Frame = +2
Query: 21 LLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYGSSFPILMIGRCIA 80
L L AL A + A ++ +GRR T++L IFF G++L G + +L+ GR +
Sbjct: 281 LFTSSLYISALVAGLGASSITRALGRRTTMILGGIFFVSGALLNGLAMNIAMLIAGRLLL 460
Query: 81 GFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGKLSLRLGWRI 140
GFG+G A V +Y +E++ YRG L LSI IG + NY+ K+ GWR+
Sbjct: 461 GFGIGCANQAVPIYLSEMAPYKYRGALNMCFQLSITIGIFTANMFNYYFSKILNGEGWRL 640
Query: 141 MLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKK 177
L + ++P++ +I + L +SP LV +GR +A+K
Sbjct: 641 SLGLGAVPAVVFIIGSICLPDSPNSLVTRGRHEEARK 751
>TC86605 similar to PIR|T01853|T01853 probable hexose transport protein
F9D12.17 - Arabidopsis thaliana, partial (91%)
Length = 3111
Score = 90.1 bits (222), Expect = 2e-18
Identities = 52/158 (32%), Positives = 79/158 (49%)
Frame = +2
Query: 19 VQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYGSSFPILMIGRC 78
+QL L L A + +GRR T++++ FF G L + +L++GR
Sbjct: 1532 LQLFTSSLYLAGLTVTFFASYTTRVLGRRLTMLIAGFFFIAGVSLNASAQNLLMLIVGRV 1711
Query: 79 IAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGKLSLRLGW 138
+ G G+GFA V V+ +EI+ RG L L L I +G L L NY K+ GW
Sbjct: 1712 LLGCGIGFANQAVPVFLSEIAPSRIRGALNILFQLDITLGILYANLVNYATNKIKGHWGW 1891
Query: 139 RIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAK 176
RI L + IP++ L + +V++P L+ +G LG K
Sbjct: 1892 RISLGLGGIPALLLTLGAYLVVDTPNSLIERGHLGQRK 2005
Score = 89.0 bits (219), Expect = 4e-18
Identities = 55/202 (27%), Positives = 95/202 (46%)
Frame = +3
Query: 239 LIAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLL 298
L+ +I + IFQ G+ I Y+P +F +G + L A + G + T++S + +
Sbjct: 2139 LVISIALMIFQQFTGINAIMFYAPVLFNTLGFKNDAALYSAVI-TGAINVISTIVSIYSV 2315
Query: 299 DKIGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIG 358
DK+GRR LLL + ++ S + + + I E ++++ I A
Sbjct: 2316 DKLGRRKLLLEAGVQMLLSQMVIAIVLGIKVKDHSEELSKGYAALVVVMVCIFVSAFAWS 2495
Query: 359 IGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTN 418
G + W+ +EIFPL R+ G V V +N + + +F+S+ G FF F G
Sbjct: 2496 WGPLAWLIPSEIFPLETRSAGQSVTVCVNFLFTAVIAQAFLSMLCYFKF-GIFFFFSGWI 2672
Query: 419 VLGWWFYYSFLPETKGRSLEDM 440
+ F + +PETK +E+M
Sbjct: 2673 LFMSTFVFFLVPETKNVPIEEM 2738
>TC81269 similar to GP|23504385|emb|CAC00697. putative sugar transporter
{Lycopersicon esculentum}, partial (42%)
Length = 966
Score = 88.6 bits (218), Expect = 5e-18
Identities = 62/207 (29%), Positives = 106/207 (50%), Gaps = 2/207 (0%)
Frame = +2
Query: 237 RILIAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDK-GTLLLATVGIGISQTLFTLLSC 295
R L +G+ + Q + G+ G+ Y+ IF G G+++ A I Q + T +
Sbjct: 35 RSLTIGVGLMVCQQLGGINGVCFYTSSIFDLAGFPSATGSIIYA-----ILQIVITGVGA 199
Query: 296 FLLDKIGRRILLLVSSGGVIFSMLGLCVCSAI-VENSKLGEEPLWAIIFTIIVIYIMAGF 354
L+D+ GR+ LLLVS G++ + V + V + +G P A+ T I++YI G
Sbjct: 200 ALIDRAGRKPLLLVSGSGLVAGCIFTAVAFYLKVHDVAVGAVPALAV--TGILVYI--GS 367
Query: 355 NAIGIGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLF 414
+IG+GA+ WV +EIFP+ ++ Q + ++N + + + + + GTF L+
Sbjct: 368 FSIGMGAIPWVVMSEIFPVNIKGQAGSIATLVNWF-GAWLCSYTFNFLMSWSSYGTFVLY 544
Query: 415 VGTNVLGWWFYYSFLPETKGRSLEDME 441
N L F +PETKG+SLE ++
Sbjct: 545 AAINALAILFIAVVVPETKGKSLEQLQ 625
>BQ751130 weakly similar to GP|15289796|dbj putative monosaccharide
transporter 3 {Oryza sativa (japonica cultivar-group)},
partial (7%)
Length = 795
Score = 88.2 bits (217), Expect = 6e-18
Identities = 62/194 (31%), Positives = 96/194 (48%), Gaps = 3/194 (1%)
Frame = +3
Query: 14 ISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILM---GYGSSF 70
IS+ L+ IL+ +IAG +SD+IGR++T+++ + +G ++ G G+
Sbjct: 165 ISESHQSLIVSILSCGTFFGALIAGDVSDWIGRKWTVIIGCGIYMIGVLIQMFTGMGAPL 344
Query: 71 PILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLG 130
++ GR IAG GVGF IV +Y +EI RG L + I IG LL Y
Sbjct: 345 GAIVAGRLIAGIGVGFESAIVILYMSEICPKKVRGALVAGYQFCITIGLLLAACVVYATK 524
Query: 131 KLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAE 190
+RI +AI ++ L +L L ESPR+ V +GR+ DA L + +E+E
Sbjct: 525 DRGDTGVYRIPIAIQFPWALILGGGLLLLPESPRFFVKKGRIADATAALSRLRGQPKESE 704
Query: 191 QRMKEIKNAVGIDE 204
E+ V +E
Sbjct: 705 YIQVELAEIVANEE 746
>TC87914 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_140
{Arabidopsis thaliana}, partial (39%)
Length = 1129
Score = 87.4 bits (215), Expect = 1e-17
Identities = 73/265 (27%), Positives = 120/265 (44%), Gaps = 13/265 (4%)
Frame = +3
Query: 210 IVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNICGVEGIFLYSPRIFGRMG 269
++H S+ T + G + ++F P V L +G+ I Q G+ G+ Y+P+I + G
Sbjct: 213 MIHPSE-TAAKGPSWNDLF---EPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAG 380
Query: 270 ITDKGTLLLATVGIGISQTLFTL--------LSCF-----LLDKIGRRILLLVSSGGVIF 316
+ LL+ +G+ + + F + L C L+D GRR LLL + +I
Sbjct: 381 VG----YLLSNLGLSSTSSSFLISAVTTLLMLPCIAVAMRLMDISGRRTLLLTTIPVLIV 548
Query: 317 SMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPLRLR 376
S+ L + S + LG+ A I TI V+ F +G G V + EIFP R+R
Sbjct: 549 SLFILVLGSLV----DLGDTAN-ASISTISVVVYFCSF-VMGFGPVPNILCAEIFPTRVR 710
Query: 377 AQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYYSFLPETKGRS 436
+ +C + I ++ V S + ++ LGG F L+ + W F + +PETKG
Sbjct: 711 GLCIAICALTFWICDIIVTYSLPVMLNSVGLGGVFGLYAVVCCIAWVFVFLKVPETKGMP 890
Query: 437 LEDMETIFGKNSNSEVQMKYGSNNA 461
LE + F + K+ A
Sbjct: 891 LEVIIEFFSVGAKQIDAAKHNCGQA 965
>TC84553 similar to GP|17381265|gb|AAL36051.1 AT5g18840/F17K4_90
{Arabidopsis thaliana}, partial (41%)
Length = 827
Score = 84.3 bits (207), Expect = 9e-17
Identities = 59/206 (28%), Positives = 105/206 (50%), Gaps = 1/206 (0%)
Frame = +2
Query: 237 RILIAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCF 296
R ++ +G+ + Q G+ GI Y+ F D + + T+ Q FT+L
Sbjct: 2 RSVVLGVGLMVCQQSVGIMGIGFYTSETFVA---ADFLSGKIGTIAYACMQVPFTILGAI 172
Query: 297 LLDKIGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGE-EPLWAIIFTIIVIYIMAGFN 355
L+DK GRR L+ S+ G + + + + L E P+ A+ I+IY+ A
Sbjct: 173 LMDKSGRRPLITASASGTFLGCFMTGIAFFLKDQNLLLELVPILAVAG--ILIYVAAF-- 340
Query: 356 AIGIGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFV 415
+IG+G V WV +EIFP+ ++ + V++N + VV+ + + + + GT FL+
Sbjct: 341 SIGMGPVPWVIMSEIFPIHVKGTAGSLVVLINWL-GAWVVSYTFNFFLSWSSPGTLFLYA 517
Query: 416 GTNVLGWWFYYSFLPETKGRSLEDME 441
G ++L F +PETKG++LE+++
Sbjct: 518 GCSLLTILFVAKLVPETKGKTLEEIQ 595
>TC91041 similar to GP|21689841|gb|AAM67564.1 unknown protein {Arabidopsis
thaliana}, partial (31%)
Length = 564
Score = 77.8 bits (190), Expect = 8e-15
Identities = 54/169 (31%), Positives = 85/169 (49%)
Frame = +3
Query: 155 LMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVS 214
LM+ L ESPRWL +G+ +AK +L I S +EAE + +K +V ++ +
Sbjct: 12 LMIMLPESPRWLFRKGKEEEAKALLRRIY-SPEEAEAEINTLKESVELE---------IK 161
Query: 215 KKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKG 274
+ S ++ +M + V R L A +G+ IFQ G+ + YSP I G
Sbjct: 162 ESESSDKASIIKMLKTKT--VRRGLYAGMGLQIFQQFVGINTVMYYSPAIIQLAGFASNQ 335
Query: 275 TLLLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVIFSMLGLCV 323
T LL ++ ++LS + +DK GR+ LLL S GV+ S++ L V
Sbjct: 336 TALLLSLVTSGLNAFGSILSIYFIDKTGRKKLLLFSLSGVVLSLVVLTV 482
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.327 0.144 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,727,957
Number of Sequences: 36976
Number of extensions: 222630
Number of successful extensions: 1614
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 1541
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1562
length of query: 461
length of database: 9,014,727
effective HSP length: 99
effective length of query: 362
effective length of database: 5,354,103
effective search space: 1938185286
effective search space used: 1938185286
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147178.8


