

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147178.7 + phase: 0
(379 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
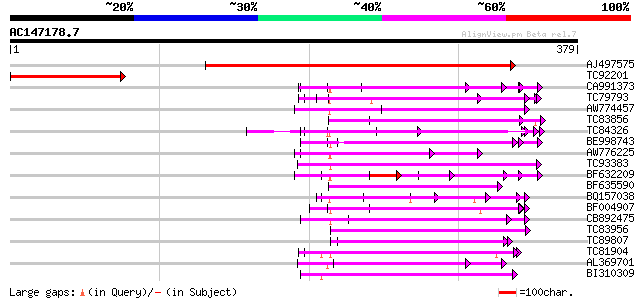
Score E
Sequences producing significant alignments: (bits) Value
AJ497575 similar to PIR|T05642|T05 hypothetical protein F20D10.2... 414 e-116
TC92201 163 1e-40
CA991373 weakly similar to PIR|D86260|D86 protein T12C24.22 [imp... 105 3e-23
TC79793 weakly similar to GP|6630464|gb|AAF19552.1| F23N19.4 {Ar... 100 7e-22
AW774457 weakly similar to GP|10176973|dbj gb|AAF19552.1~gene_id... 98 6e-21
TC83856 weakly similar to PIR|B96656|B96656 unknown protein 419... 96 2e-20
TC84326 weakly similar to PIR|F85225|F85225 hypothetical protein... 89 2e-18
BE998743 similar to GP|22128591|gb| fertility restorer-like prot... 85 5e-17
AW776225 weakly similar to GP|18461169|dbj hypothetical protein~... 76 2e-14
TC93383 similar to PIR|T47786|T47786 hypothetical protein F17J16... 74 1e-13
BF632209 weakly similar to GP|8493579|gb|A Contains a RepB PF|01... 71 6e-13
BF635590 weakly similar to PIR|D84778|D84 probable salt-inducibl... 69 3e-12
BQ157038 weakly similar to GP|4836917|gb| 80099 {Arabidopsis tha... 69 4e-12
BF004907 weakly similar to GP|8953393|emb putative protein {Arab... 68 7e-12
CB892475 weakly similar to GP|9280662|gb|A F21B7.18 {Arabidopsis... 67 1e-11
TC83956 weakly similar to GP|15982931|gb|AAL09812.1 AT3g53700/F4... 66 3e-11
TC89807 weakly similar to PIR|B96600|B96600 protein F14J16.14 [i... 66 3e-11
TC81904 weakly similar to GP|8778410|gb|AAF79418.1| F16A14.3 {Ar... 66 3e-11
AL369701 weakly similar to PIR|E84433|E8 hypothetical protein At... 64 7e-11
BI310309 similar to GP|15810431|gb unknown protein {Arabidopsis ... 64 1e-10
>AJ497575 similar to PIR|T05642|T05 hypothetical protein F20D10.270 -
Arabidopsis thaliana, partial (46%)
Length = 622
Score = 414 bits (1063), Expect = e-116
Identities = 207/207 (100%), Positives = 207/207 (100%)
Frame = +2
Query: 132 RNVAETGRDGGQSGDSFLDKFKFDFDDKIGSQSDVEASSQLEEARAVNSSNFNQPAQESM 191
RNVAETGRDGGQSGDSFLDKFKFDFDDKIGSQSDVEASSQLEEARAVNSSNFNQPAQESM
Sbjct: 2 RNVAETGRDGGQSGDSFLDKFKFDFDDKIGSQSDVEASSQLEEARAVNSSNFNQPAQESM 181
Query: 192 PQDADAIFNKMKETGLIPNAVAMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVD 251
PQDADAIFNKMKETGLIPNAVAMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVD
Sbjct: 182 PQDADAIFNKMKETGLIPNAVAMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVD 361
Query: 252 GYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSL 311
GYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSL
Sbjct: 362 GYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSL 541
Query: 312 NVTTFVGLVDGFCKENGIEEAKGVIKT 338
NVTTFVGLVDGFCKENGIEEAKGVIKT
Sbjct: 542 NVTTFVGLVDGFCKENGIEEAKGVIKT 622
>TC92201
Length = 397
Score = 163 bits (412), Expect = 1e-40
Identities = 77/77 (100%), Positives = 77/77 (100%)
Frame = +1
Query: 1 MRHFSFNDNFRGGNIKQAANGKSDDEFFRQRGDTNFNENGGNRNEEGFDIRQRLEESSQT 60
MRHFSFNDNFRGGNIKQAANGKSDDEFFRQRGDTNFNENGGNRNEEGFDIRQRLEESSQT
Sbjct: 166 MRHFSFNDNFRGGNIKQAANGKSDDEFFRQRGDTNFNENGGNRNEEGFDIRQRLEESSQT 345
Query: 61 RVLRGQKPINQPHSNSQ 77
RVLRGQKPINQPHSNSQ
Sbjct: 346 RVLRGQKPINQPHSNSQ 396
>CA991373 weakly similar to PIR|D86260|D86 protein T12C24.22 [imported] -
Arabidopsis thaliana, partial (6%)
Length = 756
Score = 105 bits (262), Expect = 3e-23
Identities = 51/142 (35%), Positives = 84/142 (58%), Gaps = 3/142 (2%)
Frame = +2
Query: 194 DADAIFNKMKETGLIPNAV---AMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVV 250
+A +F +M +IP+ V +++DGLCK G ALKL M ++G PDI+ Y++++
Sbjct: 185 EAMNLFKEMHHKHIIPDVVTYNSLIDGLCKLGKISYALKLVDEMHDRGVPPDIITYSSIL 364
Query: 251 DGYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHS 310
D K H+ D AI + K++ I PN ++YT+LI GL K +L DA ++L G++
Sbjct: 365 DALCKNHQVDKAIALLTKLKDQGIRPNMYTYTILIDGLCKGGRLEDAHNIFEDLLVKGYN 544
Query: 311 LNVTTFVGLVDGFCKENGIEEA 332
+ V T+ ++ GFC + +EA
Sbjct: 545 ITVNTYTVMIHGFCNKGLFDEA 610
Score = 87.0 bits (214), Expect = 1e-17
Identities = 44/153 (28%), Positives = 81/153 (52%), Gaps = 3/153 (1%)
Frame = +2
Query: 195 ADAIFNKMKETGLIPNAVA---MLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVD 251
A +I M + G+ P+ + ++DG CK EA+ LF M K IPD+V Y +++D
Sbjct: 83 AKSILYTMSQRGVNPDIQSYNILIDGFCKIKKVDEAMNLFKEMHHKHIIPDVVTYNSLID 262
Query: 252 GYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSL 311
G K K A+++ +M + P+ +Y+ ++ L K ++ A+ ++ + G
Sbjct: 263 GLCKLGKISYALKLVDEMHDRGVPPDIITYSSILDALCKNHQVDKAIALLTKLKDQGIRP 442
Query: 312 NVTTFVGLVDGFCKENGIEEAKGVIKTLIGKGF 344
N+ T+ L+DG CK +E+A + + L+ KG+
Sbjct: 443 NMYTYTILIDGLCKGGRLEDAHNIFEDLLVKGY 541
Score = 79.7 bits (195), Expect = 2e-15
Identities = 39/144 (27%), Positives = 75/144 (52%)
Frame = +2
Query: 213 AMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYTKAHKADDAIRIFRKMQSN 272
+++DG C +A + M ++G PDI Y ++DG+ K K D+A+ +F++M
Sbjct: 41 SLMDGYCLVKEVNKAKSILYTMSQRGVNPDIQSYNILIDGFCKIKKVDEAMNLFKEMHHK 220
Query: 273 SISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVTTFVGLVDGFCKENGIEEA 332
I P+ +Y LI GL K K+ A++ EM + G ++ T+ ++D CK + +++A
Sbjct: 221 HIIPDVVTYNSLIDGLCKLGKISYALKLVDEMHDRGVPPDIITYSSILDALCKNHQVDKA 400
Query: 333 KGVIKTLIGKGFAFDEKAVRVFLD 356
++ L +G + + +D
Sbjct: 401 IALLTKLKDQGIRPNMYTYTILID 472
Score = 64.3 bits (155), Expect = 7e-11
Identities = 36/117 (30%), Positives = 63/117 (53%), Gaps = 3/117 (2%)
Frame = +2
Query: 195 ADAIFNKMKETGLIPNAVA---MLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVD 251
A A+ K+K+ G+ PN ++DGLCK G ++A +F + KG + YT ++
Sbjct: 398 AIALLTKLKDQGIRPNMYTYTILIDGLCKGGRLEDAHNIFEDLLVKGYNITVNTYTVMIH 577
Query: 252 GYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAG 308
G+ D+A+ + KM+ NS P+A +Y ++I+ L+ + +D E EM+ G
Sbjct: 578 GFCNKGLFDEALTLLSKMKDNSCIPDAVTYEIIIRSLFDKDE-NDKAEKLREMITRG 745
Score = 56.6 bits (135), Expect = 2e-08
Identities = 31/108 (28%), Positives = 53/108 (48%)
Frame = +2
Query: 236 EKGTIPDIVIYTAVVDGYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLH 295
++G P +V Y +++DGY + + A I M ++P+ SY +LI G K K+
Sbjct: 5 KQGIKPIVVTYCSLMDGYCLVKEVNKAKSILYTMSQRGVNPDIQSYNILIDGFCKIKKVD 184
Query: 296 DAVEFCVEMLEAGHSLNVTTFVGLVDGFCKENGIEEAKGVIKTLIGKG 343
+A+ EM +V T+ L+DG CK I A ++ + +G
Sbjct: 185 EAMNLFKEMHHKHIIPDVVTYNSLIDGLCKLGKISYALKLVDEMHDRG 328
>TC79793 weakly similar to GP|6630464|gb|AAF19552.1| F23N19.4 {Arabidopsis
thaliana}, partial (7%)
Length = 1580
Score = 100 bits (250), Expect = 7e-22
Identities = 52/165 (31%), Positives = 87/165 (52%), Gaps = 3/165 (1%)
Frame = +1
Query: 194 DADAIFNKMKETGLIPNAV---AMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVV 250
+A +F +M +IP+ V +++DGL K G AL+L M ++G P+I Y +++
Sbjct: 715 EAMNLFKEMHRKNIIPDVVTYSSLIDGLSKSGRISYALQLVDQMHDRGVPPNICTYNSIL 894
Query: 251 DGYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHS 310
D K H+ D AI + K + P+ +Y++LI+GL + KL DA + +L GH+
Sbjct: 895 DALCKTHQVDKAIALLTKFKDKGFQPDISTYSILIKGLCQSGKLEDARKVFEGLLVKGHN 1074
Query: 311 LNVTTFVGLVDGFCKENGIEEAKGVIKTLIGKGFAFDEKAVRVFL 355
LNV T+ ++ GFC E EA ++ + G D K + +
Sbjct: 1075LNVDTYTIMIQGFCVEGLFNEALALLSKMEDNGCIPDAKTYEIII 1209
Score = 84.3 bits (207), Expect = 7e-17
Identities = 43/145 (29%), Positives = 80/145 (54%), Gaps = 3/145 (2%)
Frame = +1
Query: 206 GLIPNAV---AMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYTKAHKADDA 262
G+ PN V +++DG C +A +F M + G PDI Y+ +++G+ K K D+A
Sbjct: 541 GIKPNFVTYNSLMDGYCLVKEVNKAKSIFNTMAQGGVNPDIQSYSIMINGFCKIKKFDEA 720
Query: 263 IRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVTTFVGLVDG 322
+ +F++M +I P+ +Y+ LI GL K ++ A++ +M + G N+ T+ ++D
Sbjct: 721 MNLFKEMHRKNIIPDVVTYSSLIDGLSKSGRISYALQLVDQMHDRGVPPNICTYNSILDA 900
Query: 323 FCKENGIEEAKGVIKTLIGKGFAFD 347
CK + +++A ++ KGF D
Sbjct: 901 LCKTHQVDKAIALLTKFKDKGFQPD 975
Score = 63.5 bits (153), Expect = 1e-10
Identities = 39/160 (24%), Positives = 82/160 (50%), Gaps = 3/160 (1%)
Frame = +1
Query: 198 IFNKMKETGLIPNAV---AMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYT 254
+ ++M + G+ PN ++LD LCK +A+ L ++KG PDI Y+ ++ G
Sbjct: 832 LVDQMHDRGVPPNICTYNSILDALCKTHQVDKAIALLTKFKDKGFQPDISTYSILIKGLC 1011
Query: 255 KAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVT 314
++ K +DA ++F + + N +YT++IQG ++A+ +M + G +
Sbjct: 1012QSGKLEDARKVFEGLLVKGHNLNVDTYTIMIQGFCVEGLFNEALALLSKMEDNGCIPDAK 1191
Query: 315 TFVGLVDGFCKENGIEEAKGVIKTLIGKGFAFDEKAVRVF 354
T+ ++ K++ + A+ +++ +I +G + VR F
Sbjct: 1192TYEIIILSLFKKDENDMAEKLLREMIARGLP*N*NYVRYF 1311
Score = 60.1 bits (144), Expect = 1e-09
Identities = 35/105 (33%), Positives = 56/105 (53%), Gaps = 2/105 (1%)
Frame = +2
Query: 214 MLDGLCKDGNFQEALKLFGLMREKGTI--PDIVIYTAVVDGYTKAHKADDAIRIFRKMQS 271
++ GLCK G + AL L L R G + P++V+Y ++D K ++A +F +M S
Sbjct: 152 LIHGLCKVGETRAALDL--LQRVDGNLVQPNVVMYNTIIDSMCKVKLVNEAFDLFSEMVS 325
Query: 272 NSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVTTF 316
ISP+ +Y+ LI G KL DA++ +M+ +V TF
Sbjct: 326 KGISPDVVTYSALISGFCILGKLKDAIDLFNKMILENIKPDVYTF 460
Score = 30.8 bits (68), Expect = 0.90
Identities = 16/56 (28%), Positives = 30/56 (53%), Gaps = 3/56 (5%)
Frame = +2
Query: 194 DADAIFNKMKETGLIPNAV---AMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIY 246
+A +F++M G+ P+ V A++ G C G ++A+ LF M + PD+ +
Sbjct: 293 EAFDLFSEMVSKGISPDVVTYSALISGFCILGKLKDAIDLFNKMILENIKPDVYTF 460
>AW774457 weakly similar to GP|10176973|dbj
gb|AAF19552.1~gene_id:MTG13.9~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (13%)
Length = 650
Score = 97.8 bits (242), Expect = 6e-21
Identities = 50/160 (31%), Positives = 88/160 (54%), Gaps = 3/160 (1%)
Frame = -1
Query: 191 MPQDADAIFNKMKETGLIPNAV---AMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYT 247
M +A ++FN M+ G+ P+ V +++DGLCK G A +L M + G +I Y
Sbjct: 623 MVDEALSLFNDMQFKGIAPDKVTYNSLIDGLCKSGRISYAWELVDEMHDNGQPANIFTYN 444
Query: 248 AVVDGYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEA 307
++D K H D AI + +K++ I P+ +++ +LI GL K +L +A + ++L
Sbjct: 443 CLIDALCKNHHVDQAIALVKKIKDQGIQPDMYTFNILIYGLCKVGRLKNAQDVFQDLLSK 264
Query: 308 GHSLNVTTFVGLVDGFCKENGIEEAKGVIKTLIGKGFAFD 347
G+S+N T+ +V+G CKE +EA+ ++ + G D
Sbjct: 263 GYSVNAWTYNIMVNGLCKEGLFDEAEALLSKMDDNGIIPD 144
Score = 62.4 bits (150), Expect = 3e-10
Identities = 30/99 (30%), Positives = 54/99 (54%)
Frame = -1
Query: 249 VVDGYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAG 308
+++G+ K D+A+ +F MQ I+P+ +Y LI GL K ++ A E EM + G
Sbjct: 650 MINGFCKIKMVDEALSLFNDMQFKGIAPDKVTYNSLIDGLCKSGRISYAWELVDEMHDNG 471
Query: 309 HSLNVTTFVGLVDGFCKENGIEEAKGVIKTLIGKGFAFD 347
N+ T+ L+D CK + +++A ++K + +G D
Sbjct: 470 QPANIFTYNCLIDALCKNHHVDQAIALVKKIKDQGIQPD 354
>TC83856 weakly similar to PIR|B96656|B96656 unknown protein 41955-40111
[imported] - Arabidopsis thaliana, partial (12%)
Length = 662
Score = 96.3 bits (238), Expect = 2e-20
Identities = 49/148 (33%), Positives = 80/148 (53%), Gaps = 3/148 (2%)
Frame = +2
Query: 214 MLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYTKAHKADDAIRIFRKMQSNS 273
++D CK G ALKL M ++G P+IV Y++++D K H+ D A+ + K++
Sbjct: 98 LVDVFCKSGKISYALKLVDEMHDRGQPPNIVTYSSILDALCKTHRVDKAVALLTKLKDQG 277
Query: 274 ISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVTTFVGLVDGFCKENGIEEAK 333
I PN +YT+LI GL KL DA ++L G+ + V T++ + GFCK+ +EA
Sbjct: 278 IRPNMHTYTILIDGLCTSGKLEDARNIFEDLLVKGYDITVVTYIVMFYGFCKKGLFDEAS 457
Query: 334 GVIKTLIGKGFAFDEKA---VRVFLDKK 358
++ + G D K +++ L KK
Sbjct: 458 ALLSKMEENGCIPDAKTYELIKLSLFKK 541
Score = 60.1 bits (144), Expect = 1e-09
Identities = 27/104 (25%), Positives = 55/104 (51%)
Frame = +2
Query: 241 PDIVIYTAVVDGYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEF 300
PD+ + +VD + K+ K A+++ +M PN +Y+ ++ L K ++ AV
Sbjct: 74 PDVYTFNILVDVFCKSGKISYALKLVDEMHDRGQPPNIVTYSSILDALCKTHRVDKAVAL 253
Query: 301 CVEMLEAGHSLNVTTFVGLVDGFCKENGIEEAKGVIKTLIGKGF 344
++ + G N+ T+ L+DG C +E+A+ + + L+ KG+
Sbjct: 254 LTKLKDQGIRPNMHTYTILIDGLCTSGKLEDARNIFEDLLVKGY 385
Score = 35.8 bits (81), Expect = 0.028
Identities = 21/60 (35%), Positives = 26/60 (43%), Gaps = 3/60 (5%)
Frame = +2
Query: 193 QDADAIFNKMKETGL---IPNAVAMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAV 249
+DA IF + G + + M G CK G F EA L M E G IPD Y +
Sbjct: 341 EDARNIFEDLLVKGYDITVVTYIVMFYGFCKKGLFDEASALLSKMEENGCIPDAKTYELI 520
>TC84326 weakly similar to PIR|F85225|F85225 hypothetical protein AT4g19900
[imported] - Arabidopsis thaliana, partial (18%)
Length = 756
Score = 89.4 bits (220), Expect = 2e-18
Identities = 48/162 (29%), Positives = 86/162 (52%), Gaps = 3/162 (1%)
Frame = +1
Query: 195 ADAIFNKMKETGLIPNA---VAMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVD 251
A+ + ++MKE GL+PN ++DG CK GNF+ A L LM +G P++ Y A+V+
Sbjct: 49 AEMLLSRMKEQGLVPNTNTYTTLIDGHCKAGNFERAYDLMNLMSSEGFSPNLCTYNAIVN 228
Query: 252 GYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSL 311
G K + +A ++ N + P+ F+Y +L+ K + A+ +ML+ G
Sbjct: 229 GLCKRGRVQEAYKMLEDGFQNGLKPDKFTYNILMSEHCKQENIRQALALFNKMLKIGIQP 408
Query: 312 NVTTFVGLVDGFCKENGIEEAKGVIKTLIGKGFAFDEKAVRV 353
++ ++ L+ FC+EN ++E++ F E+AVR+
Sbjct: 409 DIHSYTTLIAVFCRENRMKESE-----------MFFEEAVRI 501
Score = 69.3 bits (168), Expect = 2e-12
Identities = 36/135 (26%), Positives = 65/135 (47%)
Frame = +1
Query: 213 AMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYTKAHKADDAIRIFRKMQSN 272
AM+ G C++ A L M+E+G +P+ YT ++DG+ KA + A + M S
Sbjct: 7 AMIRGYCREDKLNRAEMLLSRMKEQGLVPNTNTYTTLIDGHCKAGNFERAYDLMNLMSSE 186
Query: 273 SISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVTTFVGLVDGFCKENGIEEA 332
SPN +Y ++ GL K ++ +A + + + G + T+ L+ CK+ I +A
Sbjct: 187 GFSPNLCTYNAIVNGLCKRGRVQEAYKMLEDGFQNGLKPDKFTYNILMSEHCKQENIRQA 366
Query: 333 KGVIKTLIGKGFAFD 347
+ ++ G D
Sbjct: 367 LALFNKMLKIGIQPD 411
Score = 65.9 bits (159), Expect = 3e-11
Identities = 42/151 (27%), Positives = 67/151 (43%), Gaps = 3/151 (1%)
Frame = +1
Query: 198 IFNKMKETGLIPNAV---AMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYT 254
+ N M G PN A+++GLCK G QEA K+ + G PD Y ++ +
Sbjct: 163 LMNLMSSEGFSPNLCTYNAIVNGLCKRGRVQEAYKMLEDGFQNGLKPDKFTYNILMSEHC 342
Query: 255 KAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVT 314
K A+ +F KM I P+ SYT LI + +++ ++ F E + G
Sbjct: 343 KQENIRQALALFNKMLKIGIQPDIHSYTTLIAVFCRENRMKESEMFFEEAVRIGIIPTNK 522
Query: 315 TFVGLVDGFCKENGIEEAKGVIKTLIGKGFA 345
T+ ++ G+C+E + A L G A
Sbjct: 523 TYTSMICGYCREGNLTLAMKFFHRLSDHGCA 615
Score = 57.0 bits (136), Expect = 1e-08
Identities = 30/112 (26%), Positives = 53/112 (46%)
Frame = +1
Query: 246 YTAVVDGYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEML 305
YTA++ GY + K + A + +M+ + PN +YT LI G K A + M
Sbjct: 1 YTAMIRGYCREDKLNRAEMLLSRMKEQGLVPNTNTYTTLIDGHCKAGNFERAYDLMNLMS 180
Query: 306 EAGHSLNVTTFVGLVDGFCKENGIEEAKGVIKTLIGKGFAFDEKAVRVFLDK 357
G S N+ T+ +V+G CK ++EA +++ G D+ + + +
Sbjct: 181 SEGFSPNLCTYNAIVNGLCKRGRVQEAYKMLEDGFQNGLKPDKFTYNILMSE 336
Score = 51.2 bits (121), Expect = 6e-07
Identities = 30/121 (24%), Positives = 56/121 (45%), Gaps = 3/121 (2%)
Frame = +1
Query: 159 KIGSQSDVEASSQLEEARAVNSSNFNQPAQESMPQDADAIFNKMKETGLIPNA---VAML 215
KIG Q D+ + + L +E+ ++++ F + G+IP +M+
Sbjct: 391 KIGIQPDIHSYTTLIAVFC----------RENRMKESEMFFEEAVRIGIIPTNKTYTSMI 540
Query: 216 DGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYTKAHKADDAIRIFRKMQSNSIS 275
G C++GN A+K F + + G P+ + Y A++ G K K D+A ++ M +
Sbjct: 541 CGYCREGNLTLAMKFFHRLSDHGCAPESITYGAIISGLCKQSKRDEARSLYDSMIEKGLV 720
Query: 276 P 276
P
Sbjct: 721 P 723
>BE998743 similar to GP|22128591|gb| fertility restorer-like protein {Petunia
x hybrida}, partial (7%)
Length = 829
Score = 84.7 bits (208), Expect = 5e-17
Identities = 45/148 (30%), Positives = 82/148 (55%), Gaps = 3/148 (2%)
Frame = +2
Query: 195 ADAIFNKMKETGLIPNAVA---MLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVD 251
A + + + G+ P+A + M++G C+ A KL M G PDI Y++++D
Sbjct: 233 AKHVLSTIARMGVAPDAQSYNIMVNGFCR---ISHAWKLVDEMHVNGQPPDIFTYSSLID 403
Query: 252 GYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSL 311
K + D AI + +K++ I PN ++Y +LI GL K +L +A + ++L G+SL
Sbjct: 404 ALCKNNHLDKAIALVKKIKDQGIQPNMYTYNILIDGLCKGGRLKNAQDVFQDLLTKGYSL 583
Query: 312 NVTTFVGLVDGFCKENGIEEAKGVIKTL 339
N+ T+ L++G CKE ++A+ ++ +
Sbjct: 584 NIRTYNILINGLCKEGLFDKAEALLSKM 667
Score = 64.3 bits (155), Expect = 7e-11
Identities = 33/137 (24%), Positives = 68/137 (49%)
Frame = +2
Query: 220 KDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYTKAHKADDAIRIFRKMQSNSISPNAF 279
K+GN + A +M + PD+V Y +++DGY ++ + A + + ++P+A
Sbjct: 107 KEGNVKGAKNALAMMIKGSIKPDVVTYNSLMDGYCLVNEVNKAKHVLSTIARMGVAPDAQ 286
Query: 280 SYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVTTFVGLVDGFCKENGIEEAKGVIKTL 339
SY +++ G + S V+ EM G ++ T+ L+D CK N +++A ++K +
Sbjct: 287 SYNIMVNGFCRISHAWKLVD---EMHVNGQPPDIFTYSSLIDALCKNNHLDKAIALVKKI 457
Query: 340 IGKGFAFDEKAVRVFLD 356
+G + + +D
Sbjct: 458 KDQGIQPNMYTYNILID 508
Score = 64.3 bits (155), Expect = 7e-11
Identities = 35/144 (24%), Positives = 71/144 (49%)
Frame = +2
Query: 213 AMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYTKAHKADDAIRIFRKMQSN 272
+++DG C +A + + G PD Y +V+G+ + A ++ +M N
Sbjct: 191 SLMDGYCLVNEVNKAKHVLSTIARMGVAPDAQSYNIMVNGFCRI---SHAWKLVDEMHVN 361
Query: 273 SISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVTTFVGLVDGFCKENGIEEA 332
P+ F+Y+ LI L K + L A+ ++ + G N+ T+ L+DG CK ++ A
Sbjct: 362 GQPPDIFTYSSLIDALCKNNHLDKAIALVKKIKDQGIQPNMYTYNILIDGLCKGGRLKNA 541
Query: 333 KGVIKTLIGKGFAFDEKAVRVFLD 356
+ V + L+ KG++ + + + ++
Sbjct: 542 QDVFQDLLTKGYSLNIRTYNILIN 613
Score = 58.5 bits (140), Expect = 4e-09
Identities = 30/131 (22%), Positives = 69/131 (51%)
Frame = +2
Query: 213 AMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYTKAHKADDAIRIFRKMQSN 272
+++D LCK+ + +A+ L ++++G P++ Y ++DG K + +A +F+ + +
Sbjct: 392 SLIDALCKNNHLDKAIALVKKIKDQGIQPNMYTYNILIDGLCKGGRLKNAQDVFQDLLTK 571
Query: 273 SISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVTTFVGLVDGFCKENGIEEA 332
S N +Y +LI GL K A +M + + NV T+ ++ ++ E+A
Sbjct: 572 GYSLNIRTYNILINGLCKEGLFDKAEALLSKMEDNDINPNVVTYETIIRSLFYKDYNEKA 751
Query: 333 KGVIKTLIGKG 343
+ +++ ++ +G
Sbjct: 752 EKLLREMVARG 784
Score = 33.9 bits (76), Expect = 0.11
Identities = 15/61 (24%), Positives = 31/61 (50%)
Frame = +2
Query: 214 MLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYTKAHKADDAIRIFRKMQSNS 273
+++GLCK+G F +A L M + P++V Y ++ + A ++ R+M +
Sbjct: 605 LINGLCKEGLFDKAEALLSKMEDNDINPNVVTYETIIRSLFYKDYNEKAEKLLREMVARG 784
Query: 274 I 274
+
Sbjct: 785 L 787
>AW776225 weakly similar to GP|18461169|dbj hypothetical protein~similar to
Arabidopsis thaliana chromosome 5 MCL19.15, partial
(10%)
Length = 396
Score = 75.9 bits (185), Expect = 2e-14
Identities = 35/97 (36%), Positives = 56/97 (57%), Gaps = 3/97 (3%)
Frame = +1
Query: 191 MPQDADAIFNKMKETGLIPNAV---AMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYT 247
M A +F +M + PN + +++DGLCK G AL+L LM ++G PDI+ Y+
Sbjct: 106 MVDQAMKLFKEMHHKQIFPNVITYNSLIDGLCKSGRISYALELIDLMHDRGQQPDIITYS 285
Query: 248 AVVDGYTKAHKADDAIRIFRKMQSNSISPNAFSYTVL 284
+++D K H D AI + K++ I PN ++YT+L
Sbjct: 286 SILDALCKNHLVDKAIALLIKLKDQGIRPNMYTYTIL 396
Score = 54.7 bits (130), Expect = 6e-08
Identities = 29/125 (23%), Positives = 60/125 (47%), Gaps = 3/125 (2%)
Frame = +1
Query: 195 ADAIFNKMKETGLIPNAVA---MLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVD 251
A +I N M G+ + +++G CK +A+KLF M K P+++ Y +++D
Sbjct: 13 AKSILNTMSHRGVTATVRSYNIVINGFCKIKMVDQAMKLFKEMHHKQIFPNVITYNSLID 192
Query: 252 GYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSL 311
G K+ + A+ + M P+ +Y+ ++ L K + A+ +++ + G
Sbjct: 193 GLCKSGRISYALELIDLMHDRGQQPDIITYSSILDALCKNHLVDKAIALLIKLKDQGIRP 372
Query: 312 NVTTF 316
N+ T+
Sbjct: 373 NMYTY 387
Score = 38.1 bits (87), Expect = 0.006
Identities = 22/79 (27%), Positives = 34/79 (42%)
Frame = +1
Query: 265 IFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVTTFVGLVDGFC 324
I M ++ SY ++I G K + A++ EM NV T+ L+DG C
Sbjct: 22 ILNTMSHRGVTATVRSYNIVINGFCKIKMVDQAMKLFKEMHHKQIFPNVITYNSLIDGLC 201
Query: 325 KENGIEEAKGVIKTLIGKG 343
K I A +I + +G
Sbjct: 202 KSGRISYALELIDLMHDRG 258
>TC93383 similar to PIR|T47786|T47786 hypothetical protein F17J16.90 -
Arabidopsis thaliana, partial (43%)
Length = 1038
Score = 73.6 bits (179), Expect = 1e-13
Identities = 45/166 (27%), Positives = 76/166 (45%), Gaps = 3/166 (1%)
Frame = +1
Query: 193 QDADAIFNKMKETGLIPNAVA---MLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAV 249
++A A+F +M + G+ P A +LD G ++A +F MR +PD+ YT +
Sbjct: 151 EEALAVFEEMLDAGVRPTRKAYNILLDAFSISGMVEQARIVFKSMRRDKYMPDLCSYTTM 330
Query: 250 VDGYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGH 309
+ Y A + A + F+++ + PN +Y LI+G K + + +E EML G
Sbjct: 331 LSAYVNAPDMEGAEKFFKRLIQDGFEPNVVTYGTLIKGYAKANDIEKVMEKYEEMLGRGI 510
Query: 310 SLNVTTFVGLVDGFCKENGIEEAKGVIKTLIGKGFAFDEKAVRVFL 355
N T ++D K + A K + G D+KA + L
Sbjct: 511 KANQTILTTIMDAHGKNGDFDSAVNWFKEMALNGLLPDQKAKNILL 648
>BF632209 weakly similar to GP|8493579|gb|A Contains a RepB PF|01051 protein
domain and multiple PPR PF|01535 repeats. EST
gb|AA728420 comes, partial (13%)
Length = 508
Score = 71.2 bits (173), Expect = 6e-13
Identities = 37/110 (33%), Positives = 62/110 (55%), Gaps = 3/110 (2%)
Frame = +1
Query: 191 MPQDADAIFNKMKETGLIPNAV---AMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYT 247
M A ++F +M+ + P+ V +++DGLCK G A +L MR+ G DI+ Y
Sbjct: 160 MVDKALSLFYEMRCRRIAPDTVTYNSLIDGLCKSGRISYAWELVDEMRDSGQPADIITYN 339
Query: 248 AVVDGYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDA 297
+++D K H D AI + +K++ I + ++Y +LI GL K +L+DA
Sbjct: 340 SLIDALCKNHHVDKAIALVKKIKDQGIQLDMYTYNILIDGLCKQGRLNDA 489
Score = 69.3 bits (168), Expect = 2e-12
Identities = 32/116 (27%), Positives = 62/116 (52%)
Frame = +1
Query: 241 PDIVIYTAVVDGYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEF 300
P+ Y+ V++G+ K D A+ +F +M+ I+P+ +Y LI GL K ++ A E
Sbjct: 109 PNARSYSIVINGFCKIKMVDKALSLFYEMRCRRIAPDTVTYNSLIDGLCKSGRISYAWEL 288
Query: 301 CVEMLEAGHSLNVTTFVGLVDGFCKENGIEEAKGVIKTLIGKGFAFDEKAVRVFLD 356
EM ++G ++ T+ L+D CK + +++A ++K + +G D + +D
Sbjct: 289 VDEMRDSGQPADIITYNSLIDALCKNHHVDKAIALVKKIKDQGIQLDMYTYNILID 456
Score = 60.5 bits (145), Expect = 1e-09
Identities = 36/128 (28%), Positives = 66/128 (51%), Gaps = 3/128 (2%)
Frame = +1
Query: 209 PNAVA---MLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYTKAHKADDAIRI 265
PNA + +++G CK +AL LF MR + PD V Y +++DG K+ + A +
Sbjct: 109 PNARSYSIVINGFCKIKMVDKALSLFYEMRCRRIAPDTVTYNSLIDGLCKSGRISYAWEL 288
Query: 266 FRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVTTFVGLVDGFCK 325
+M+ + + +Y LI L K + A+ ++ + G L++ T+ L+DG CK
Sbjct: 289 VDEMRDSGQPADIITYNSLIDALCKNHHVDKAIALVKKIKDQGIQLDMYTYNILIDGLCK 468
Query: 326 ENGIEEAK 333
+ + +A+
Sbjct: 469 QGRLNDAQ 492
Score = 37.7 bits (86), Expect(2) = 2e-04
Identities = 20/70 (28%), Positives = 34/70 (48%)
Frame = +1
Query: 274 ISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVTTFVGLVDGFCKENGIEEAK 333
++PNA SY+++I G K + A+ EM + + T+ L+DG CK I A
Sbjct: 103 VAPNARSYSIVINGFCKIKMVDKALSLFYEMRCRRIAPDTVTYNSLIDGLCKSGRISYAW 282
Query: 334 GVIKTLIGKG 343
++ + G
Sbjct: 283 ELVDEMRDSG 312
Score = 36.6 bits (83), Expect = 0.016
Identities = 18/72 (25%), Positives = 42/72 (58%), Gaps = 3/72 (4%)
Frame = +1
Query: 198 IFNKMKETGLIPNAV---AMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYT 254
+ ++M+++G + + +++D LCK+ + +A+ L ++++G D+ Y ++DG
Sbjct: 286 LVDEMRDSGQPADIITYNSLIDALCKNHHVDKAIALVKKIKDQGIQLDMYTYNILIDGLC 465
Query: 255 KAHKADDAIRIF 266
K + +DA IF
Sbjct: 466 KQGRLNDAQVIF 501
Score = 24.6 bits (52), Expect(2) = 2e-04
Identities = 8/22 (36%), Positives = 16/22 (72%)
Frame = +2
Query: 241 PDIVIYTAVVDGYTKAHKADDA 262
PD+V Y++++DGY ++ + A
Sbjct: 2 PDVVTYSSLMDGYCLVNEVNKA 67
>BF635590 weakly similar to PIR|D84778|D84 probable salt-inducible protein
[imported] - Arabidopsis thaliana, partial (25%)
Length = 465
Score = 68.9 bits (167), Expect = 3e-12
Identities = 33/116 (28%), Positives = 59/116 (50%)
Frame = +3
Query: 214 MLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYTKAHKADDAIRIFRKMQSNS 273
++ G K G F A + + M + PD+ + ++ GY + K A+ +F +M+
Sbjct: 117 LIHGFVKSGRFDRAFEFYNQMVKDRIKPDVFTFNILISGYCRDFKFGFALEMFDEMRKMG 296
Query: 274 ISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVTTFVGLVDGFCKENGI 329
PN ++ LI+GL++ ++ + + EM+E G L+ T LVDG CKE +
Sbjct: 297 CHPNVVTFNTLIKGLFRECRVDEGIGMVYEMIELGCQLSDVTCEILVDGLCKEGRV 464
>BQ157038 weakly similar to GP|4836917|gb| 80099 {Arabidopsis thaliana},
partial (4%)
Length = 653
Score = 68.6 bits (166), Expect = 4e-12
Identities = 38/122 (31%), Positives = 59/122 (48%), Gaps = 11/122 (9%)
Frame = +1
Query: 237 KGTIPDIVIYTAVVDGYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHD 296
KG +PD+V YTA++DG+ K + A + ++M ++PN + T LI GL K K +
Sbjct: 10 KGLVPDVVTYTALIDGHCKVENSKVAFELHKEMMEAGLTPNVVTVTSLIDGLLKEGKTYG 189
Query: 297 AVEFCVEMLEAGH-----------SLNVTTFVGLVDGFCKENGIEEAKGVIKTLIGKGFA 345
A++ +E E G S N + L+ G CK+ I +A K + GF
Sbjct: 190 AIKLFLEKTEVGSPGGKTDHSGVCSPNEVMYAALIQGLCKDGRIFKATKFFKDMRCSGFK 369
Query: 346 FD 347
D
Sbjct: 370 PD 375
Score = 63.2 bits (152), Expect = 2e-10
Identities = 41/130 (31%), Positives = 66/130 (50%), Gaps = 14/130 (10%)
Frame = +1
Query: 206 GLIPNAV---AMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYTKAHKADDA 262
GL+P+ V A++DG CK N + A +L M E G P++V T+++DG K K A
Sbjct: 13 GLVPDVVTYTALIDGHCKVENSKVAFELHKEMMEAGLTPNVVTVTSLIDGLLKEGKTYGA 192
Query: 263 IRIF-----------RKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSL 311
I++F + S SPN Y LIQGL K ++ A +F +M +G
Sbjct: 193 IKLFLEKTEVGSPGGKTDHSGVCSPNEVMYAALIQGLCKDGRIFKATKFFKDMRCSGFKP 372
Query: 312 NVTTFVGLVD 321
++ +V +++
Sbjct: 373 DMVLYVIMLE 402
Score = 44.3 bits (103), Expect = 8e-05
Identities = 26/82 (31%), Positives = 40/82 (48%), Gaps = 3/82 (3%)
Frame = +1
Query: 209 PNAV---AMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYTKAHKADDAIRI 265
PN V A++ GLCKDG +A K F MR G PD+V+Y +++ + + D + +
Sbjct: 265 PNEVMYAALIQGLCKDGRIFKATKFFKDMRCSGFKPDMVLYVIMLEAHFRFKHMFDVMML 444
Query: 266 FRKMQSNSISPNAFSYTVLIQG 287
M + N VL +G
Sbjct: 445 HADMLKTGVLRNTSVCRVLTRG 510
Score = 42.7 bits (99), Expect = 2e-04
Identities = 27/74 (36%), Positives = 35/74 (46%)
Frame = +1
Query: 269 MQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVTTFVGLVDGFCKENG 328
M + P+ +YT LI G K A E EM+EAG + NV T L+DG KE
Sbjct: 1 MVIKGLVPDVVTYTALIDGHCKVENSKVAFELHKEMMEAGLTPNVVTVTSLIDGLLKEG- 177
Query: 329 IEEAKGVIKTLIGK 342
+ G IK + K
Sbjct: 178 --KTYGAIKLFLEK 213
>BF004907 weakly similar to GP|8953393|emb putative protein {Arabidopsis
thaliana}, partial (34%)
Length = 665
Score = 67.8 bits (164), Expect = 7e-12
Identities = 40/150 (26%), Positives = 74/150 (48%), Gaps = 3/150 (2%)
Frame = +3
Query: 201 KMKETGLIPNAVA---MLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYTKAH 257
+M + G+ PNA+ ++D L + G F+EAL + P + Y ++V G+ KA
Sbjct: 102 EMTKEGIKPNAIVYNPIIDALAEAGRFKEALGMMERFHVLQIGPTLSTYNSLVKGFCKAG 281
Query: 258 KADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVTTFV 317
+ A +I +KM S P +Y + +C K+ + + +M+E+GH+ + T+
Sbjct: 282 DIEGASKILKKMISRGFLPIPTTYNYFFRYFSRCGKVDEGMNLYTKMIESGHNPDRLTYH 461
Query: 318 GLVDGFCKENGIEEAKGVIKTLIGKGFAFD 347
++ C+E +E A V + KG+ D
Sbjct: 462 LVLKMLCEEEKLELAVQVSMEMRHKGYDMD 551
Score = 64.3 bits (155), Expect = 7e-11
Identities = 34/108 (31%), Positives = 56/108 (51%), Gaps = 4/108 (3%)
Frame = +3
Query: 241 PDIVIYTAVVDGYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEF 300
P +V Y +V+GY + + + A+ + +M I PNA Y +I L + + +A
Sbjct: 21 PSVVTYGTLVEGYCRMRRVEKALEMVGEMTKEGIKPNAIVYNPIIDALAEAGRFKEA--- 191
Query: 301 CVEMLEAGHSLNV----TTFVGLVDGFCKENGIEEAKGVIKTLIGKGF 344
+ M+E H L + +T+ LV GFCK IE A ++K +I +GF
Sbjct: 192 -LGMMERFHVLQIGPTLSTYNSLVKGFCKAGDIEGASKILKKMISRGF 332
Score = 62.0 bits (149), Expect = 4e-10
Identities = 33/131 (25%), Positives = 68/131 (51%)
Frame = +3
Query: 213 AMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYTKAHKADDAIRIFRKMQSN 272
+++ G CK G+ + A K+ M +G +P Y +++ K D+ + ++ KM +
Sbjct: 252 SLVKGFCKAGDIEGASKILKKMISRGFLPIPTTYNYFFRYFSRCGKVDEGMNLYTKMIES 431
Query: 273 SISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVTTFVGLVDGFCKENGIEEA 332
+P+ +Y ++++ L + KL AV+ +EM G+ +++ T L CK + +EEA
Sbjct: 432 GHNPDRLTYHLVLKMLCEEEKLELAVQVSMEMRHKGYDMDLATSTMLTHLLCKMHKLEEA 611
Query: 333 KGVIKTLIGKG 343
+ +I +G
Sbjct: 612 FAEFEDMIRRG 644
>CB892475 weakly similar to GP|9280662|gb|A F21B7.18 {Arabidopsis thaliana},
partial (5%)
Length = 527
Score = 67.0 bits (162), Expect = 1e-11
Identities = 37/144 (25%), Positives = 74/144 (50%), Gaps = 3/144 (2%)
Frame = +1
Query: 195 ADAIFNKMKETGLIPNAVA---MLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVD 251
A +FN MKET P+ ++ +L+G CK G +EAL +++ G + Y ++++
Sbjct: 1 AHKLFNDMKETSYPPDMISCNVVLNGFCKMGRLEEALSFVWMIKNDGFSLNRNSYASLIN 180
Query: 252 GYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSL 311
+ KA + +A + KM I P+ Y ++I+GL K ++ +A + EM + G +
Sbjct: 181 AFFKARRYREAHACYTKMFKEGIVPDVVLYAIMIRGLSKEGRVGEAAKMLEEMTQIGLTP 360
Query: 312 NVTTFVGLVDGFCKENGIEEAKGV 335
+ + ++ G C + + A+ +
Sbjct: 361 DSYCYNAVIQGLCDVDLFDRAQSL 432
Score = 50.4 bits (119), Expect = 1e-06
Identities = 30/121 (24%), Positives = 58/121 (47%)
Frame = +1
Query: 227 ALKLFGLMREKGTIPDIVIYTAVVDGYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQ 286
A KLF M+E PD++ V++G+ K + ++A+ ++++ S N SY LI
Sbjct: 1 AHKLFNDMKETSYPPDMISCNVVLNGFCKMGRLEEALSFVWMIKNDGFSLNRNSYASLIN 180
Query: 287 GLYKCSKLHDAVEFCVEMLEAGHSLNVTTFVGLVDGFCKENGIEEAKGVIKTLIGKGFAF 346
+K + +A +M + G +V + ++ G KE + EA +++ + G
Sbjct: 181 AFFKARRYREAHACYTKMFKEGIVPDVVLYAIMIRGLSKEGRVGEAAKMLEEMTQIGLTP 360
Query: 347 D 347
D
Sbjct: 361 D 363
>TC83956 weakly similar to GP|15982931|gb|AAL09812.1 AT3g53700/F4P12_400
{Arabidopsis thaliana}, partial (18%)
Length = 762
Score = 65.9 bits (159), Expect = 3e-11
Identities = 37/134 (27%), Positives = 69/134 (50%)
Frame = +1
Query: 215 LDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYTKAHKADDAIRIFRKMQSNSI 274
L+ L +D + L M +G + D+ + ++ KAH+ AI + +M ++ +
Sbjct: 313 LNALVEDNKLKLVEMLHSKMVNEGIVLDVSTFNVLIKALCKAHQLRPAILMLEEMANHGL 492
Query: 275 SPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVTTFVGLVDGFCKENGIEEAKG 334
P+ ++T L+QG + L+ A++ +ML G L + LV+GFCKE +EEA
Sbjct: 493 KPDEITFTTLMQGFIEEGDLNGALKMKKQMLGYGCLLTNVSVKVLVNGFCKEGRVEEALR 672
Query: 335 VIKTLIGKGFAFDE 348
+ + +GF+ D+
Sbjct: 673 FVLEVSEEGFSPDQ 714
Score = 32.3 bits (72), Expect = 0.31
Identities = 14/45 (31%), Positives = 27/45 (59%)
Frame = +1
Query: 214 MLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYTKAHK 258
+++G CK+G +EAL+ + E+G PD + ++V+G + K
Sbjct: 625 LVNGFCKEGRVEEALRFVLEVSEEGFSPDQGTFNSLVNGVCRIWK 759
>TC89807 weakly similar to PIR|B96600|B96600 protein F14J16.14 [imported] -
Arabidopsis thaliana, partial (53%)
Length = 1241
Score = 65.9 bits (159), Expect = 3e-11
Identities = 32/122 (26%), Positives = 63/122 (51%)
Frame = +1
Query: 215 LDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYTKAHKADDAIRIFRKMQSNSI 274
+ L + G+F A+ + M + G D++ + ++DG + +D +++ K+ ++
Sbjct: 496 IKALLEKGSFDSAVSVLEEMEKDGVESDLITFNTLLDGLYSKGRFEDGEKLWEKLGEKNV 675
Query: 275 SPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVTTFVGLVDGFCKENGIEEAKG 334
PN +Y + GL + +AVEF EM + G ++ +F L+ GF E ++EAK
Sbjct: 676 VPNIRTYNARLLGLAVAKRAGEAVEFYEEMEKKGVKPDLFSFNALIKGFANEGNLDEAKN 855
Query: 335 VI 336
V+
Sbjct: 856 VV 861
Score = 43.5 bits (101), Expect = 1e-04
Identities = 29/115 (25%), Positives = 52/115 (45%), Gaps = 1/115 (0%)
Frame = +1
Query: 220 KDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYTKAHKADDAIRIFRKMQSN-SISPNA 278
K + A KLF M ++ ++ A++ Y + + D R+F+K+ S+ P+
Sbjct: 298 KSNMHRHAQKLFDEMPQRNCERSVLSLNALLAAYLHSKQYDVVERLFKKLPVQLSVKPDL 477
Query: 279 FSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVTTFVGLVDGFCKENGIEEAK 333
SY I+ L + AV EM + G ++ TF L+DG + E+ +
Sbjct: 478 VSYNTYIKALLEKGSFDSAVSVLEEMEKDGVESDLITFNTLLDGLYSKGRFEDGE 642
>TC81904 weakly similar to GP|8778410|gb|AAF79418.1| F16A14.3 {Arabidopsis
thaliana}, partial (7%)
Length = 978
Score = 65.9 bits (159), Expect = 3e-11
Identities = 39/156 (25%), Positives = 80/156 (51%), Gaps = 9/156 (5%)
Frame = +1
Query: 194 DADAIFNKMKETGL---IPNAVAMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVV 250
DA + +++K L + + +++G G EA LF M E+G PD+V Y +
Sbjct: 331 DAVGMLDELKSMQLDVDMKHYTTLINGYFLQGKPIEAQSLFKEMEERGFKPDVVAYNVLA 510
Query: 251 DGYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHS 310
G+ + +A+ + M+S + PN+ ++ ++I+GL K+ +A EF +
Sbjct: 511 AGFFRNRTDFEAMDLLNYMESQGVEPNSTTHKIIIEGLCSAGKVEEAEEFFNWLKGESVE 690
Query: 311 LNVTTFVGLVDGFC------KENGIEEAKGVIKTLI 340
++V + LV+G+C K + ++EA +++T++
Sbjct: 691 ISVEIYTALVNGYCEAALIEKSHELKEAFILLRTML 798
Score = 65.5 bits (158), Expect = 3e-11
Identities = 39/148 (26%), Positives = 71/148 (47%), Gaps = 3/148 (2%)
Frame = +1
Query: 198 IFNKMKETGLIPNAVA---MLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYT 254
+F + KE+GL + A + D LCK G +A+ + ++ D+ YT +++GY
Sbjct: 238 MFEEFKESGLFIDRKAYNILFDALCKLGKVDDAVGMLDELKSMQLDVDMKHYTTLINGYF 417
Query: 255 KAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVT 314
K +A +F++M+ P+ +Y VL G ++ +A++ M G N T
Sbjct: 418 LQGKPIEAQSLFKEMEERGFKPDVVAYNVLAAGFFRNRTDFEAMDLLNYMESQGVEPNST 597
Query: 315 TFVGLVDGFCKENGIEEAKGVIKTLIGK 342
T +++G C +EEA+ L G+
Sbjct: 598 THKIIIEGLCSAGKVEEAEEFFNWLKGE 681
Score = 32.0 bits (71), Expect = 0.40
Identities = 19/58 (32%), Positives = 27/58 (45%), Gaps = 3/58 (5%)
Frame = +1
Query: 193 QDADAIFNKMKETGLIPNAVA---MLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYT 247
++A + M E + P+ V + LC +GN + A LF L G PD V YT
Sbjct: 766 KEAFILLRTMLEMNMKPSKVMYSKIFTALCCNGNMEGAHTLFNLFXHTGFTPDAVTYT 939
>AL369701 weakly similar to PIR|E84433|E8 hypothetical protein At2g02150
[imported] - Arabidopsis thaliana, partial (11%)
Length = 463
Score = 64.3 bits (155), Expect = 7e-11
Identities = 34/143 (23%), Positives = 73/143 (50%), Gaps = 3/143 (2%)
Frame = +1
Query: 193 QDADAIFNKMKETGLIPNA---VAMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAV 249
++A+ +F + E G+ N +++ G K ++A+ + M +K D +Y
Sbjct: 1 KEAEELFRALLEAGVALNLQIYTSLIHGYMKAKMMEKAMDILKEMNKKNFKLDSPLYGTK 180
Query: 250 VDGYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGH 309
+ G +K +++ + R+M+ + ++ N++ YT L+ +K K+ +AV EM + G
Sbjct: 181 IWGLCSQNKIEESEAVMREMKDHGLTANSYIYTSLMDAYFKVGKITEAVNLLQEMQKFGI 360
Query: 310 SLNVTTFVGLVDGFCKENGIEEA 332
T+ L+DG CK+ +++A
Sbjct: 361 ETTAVTYGVLIDGLCKKGLVQQA 429
Score = 54.7 bits (130), Expect = 6e-08
Identities = 29/92 (31%), Positives = 47/92 (50%)
Frame = +1
Query: 217 GLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVDGYTKAHKADDAIRIFRKMQSNSISP 276
GLC +E+ + M++ G + IYT+++D Y K K +A+ + ++MQ I
Sbjct: 187 GLCSQNKIEESEAVMREMKDHGLTANSYIYTSLMDAYFKVGKITEAVNLLQEMQKFGIET 366
Query: 277 NAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAG 308
A +Y VLI GL K + AV + M + G
Sbjct: 367 TAVTYGVLIDGLCKKGLVQQAVSYFDCMTKTG 462
Score = 40.4 bits (93), Expect = 0.001
Identities = 22/83 (26%), Positives = 40/83 (47%)
Frame = +1
Query: 261 DAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSLNVTTFVGLV 320
+A +FR + ++ N YT LI G K + A++ EM + L+ + +
Sbjct: 4 EAEELFRALLEAGVALNLQIYTSLIHGYMKAKMMEKAMDILKEMNKKNFKLDSPLYGTKI 183
Query: 321 DGFCKENGIEEAKGVIKTLIGKG 343
G C +N IEE++ V++ + G
Sbjct: 184 WGLCSQNKIEESEAVMREMKDHG 252
Score = 30.4 bits (67), Expect = 1.2
Identities = 16/48 (33%), Positives = 27/48 (55%), Gaps = 3/48 (6%)
Frame = +1
Query: 194 DADAIFNKMKETGLIPNAVA---MLDGLCKDGNFQEALKLFGLMREKG 238
+A + +M++ G+ AV ++DGLCK G Q+A+ F M + G
Sbjct: 319 EAVNLLQEMQKFGIETTAVTYGVLIDGLCKKGLVQQAVSYFDCMTKTG 462
>BI310309 similar to GP|15810431|gb unknown protein {Arabidopsis thaliana},
partial (37%)
Length = 634
Score = 63.9 bits (154), Expect = 1e-10
Identities = 41/148 (27%), Positives = 73/148 (48%), Gaps = 3/148 (2%)
Frame = +1
Query: 195 ADAIFNKMKETGL---IPNAVAMLDGLCKDGNFQEALKLFGLMREKGTIPDIVIYTAVVD 251
A+A+ +M+E G+ I M+DG GN ++ L +F ++E G P IV Y +++
Sbjct: 100 AEALVREMEEQGIDAPIDIYHTMMDGYTMIGNEEKCLIVFERLKECGFSPSIVSYGCLIN 279
Query: 252 GYTKAHKADDAIRIFRKMQSNSISPNAFSYTVLIQGLYKCSKLHDAVEFCVEMLEAGHSL 311
YTK K A+ I R M++ I N +Y++L G K +A ++ + G
Sbjct: 280 LYTKIGKVSKALEISRVMKTVGIKHNMKTYSMLFNGFVKLKDWANAFSVFEDITKDGLKP 459
Query: 312 NVTTFVGLVDGFCKENGIEEAKGVIKTL 339
+V + +V FC ++ A ++K +
Sbjct: 460 DVILYNNIVKAFCGMGNMDRAICIVKQM 543
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.136 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,141,154
Number of Sequences: 36976
Number of extensions: 105456
Number of successful extensions: 708
Number of sequences better than 10.0: 107
Number of HSP's better than 10.0 without gapping: 573
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 676
length of query: 379
length of database: 9,014,727
effective HSP length: 98
effective length of query: 281
effective length of database: 5,391,079
effective search space: 1514893199
effective search space used: 1514893199
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC147178.7


