

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147178.14 + phase: 0
(465 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
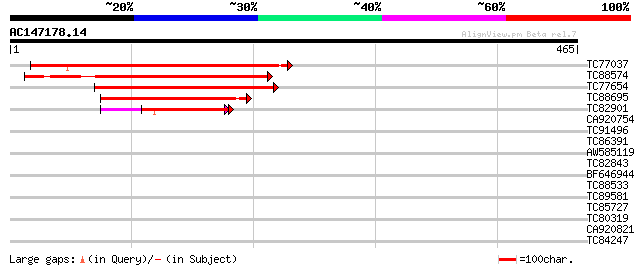
Score E
Sequences producing significant alignments: (bits) Value
TC77037 similar to PIR|B84652|B84652 hypothetical protein At2g25... 280 8e-76
TC88574 weakly similar to GP|22137236|gb|AAM91463.1 At2g36630/F1... 218 3e-57
TC77654 similar to GP|22137236|gb|AAM91463.1 At2g36630/F1O11.26 ... 216 1e-56
TC88695 similar to GP|13272465|gb|AAK17171.1 unknown protein {Ar... 100 9e-22
TC82901 similar to GP|22137236|gb|AAM91463.1 At2g36630/F1O11.26 ... 75 4e-14
CA920754 similar to PIR|T35781|T35 hypothetical protein SC8A6.14... 35 0.061
TC91496 similar to GP|13272465|gb|AAK17171.1 unknown protein {Ar... 32 0.68
TC86391 similar to GP|4097571|gb|AAD09514.1| GMFP5 {Glycine max}... 31 1.2
AW585119 31 1.2
TC82843 similar to PIR|A44226|A44226 auxin-independent growth pr... 30 1.5
BF646944 similar to GP|12597808|gb| hypothetical protein {Arabid... 30 2.6
TC88533 similar to GP|12081919|dbj|BAB20862. plastidic cysteine ... 28 5.7
TC89581 similar to PIR|B96661|B96661 unknown protein 83181-8510... 28 7.5
TC85727 homologue to PIR|D84431|D84431 probable endosomal protei... 28 9.8
TC80319 similar to GP|20259261|gb|AAM14366.1 putative HhoA prote... 28 9.8
CA920821 similar to GP|16930441|gb| At2g42810/F7D19.19 {Arabidop... 28 9.8
TC84247 weakly similar to GP|14269077|gb|AAK58011.1 verticillium... 28 9.8
>TC77037 similar to PIR|B84652|B84652 hypothetical protein At2g25740
[imported] - Arabidopsis thaliana, partial (33%)
Length = 2114
Score = 280 bits (716), Expect = 8e-76
Identities = 138/219 (63%), Positives = 174/219 (79%), Gaps = 4/219 (1%)
Frame = +2
Query: 18 WLIMCILVMICNVSLAERVLKEKEPAKFV----EKETKGFLKAMVDFLWESGKSSYEPVW 73
W+++ + VS +++K + P+ V +K + FL V+FLW+S S Y+ VW
Sbjct: 419 WMMLLLFGSFLIVSGERKLVKIQLPSFNVTTQPQKHEQSFLTKAVNFLWKSDGSGYQHVW 598
Query: 74 PEMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIMGA 133
PEM+F W+I++GS IGF GAA GSVGGVGGGGIFVPML+LIIGFDPKSSTAISKCMIMGA
Sbjct: 599 PEMEFGWQIVLGSFIGFCGAAFGSVGGVGGGGIFVPMLSLIIGFDPKSSTAISKCMIMGA 778
Query: 134 ALSTVYYNMRLRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIILF 193
ALSTVYYN+RLR+PTLDMP+IDYDLALL QPMLMLGISIGV+ NV+F DW+VT+LLI+LF
Sbjct: 779 ALSTVYYNLRLRHPTLDMPIIDYDLALLIQPMLMLGISIGVVFNVVFPDWLVTILLIVLF 958
Query: 194 IGTSTKALIKGINTWKKETMLKKETAKQLEEEPKTGAYQ 232
+GTSTKA KG+ TW KET++KKE A++ +E +G Y+
Sbjct: 959 LGTSTKAFFKGLETWNKETIMKKEAARR-QESNGSGEYK 1072
Score = 33.1 bits (74), Expect = 0.23
Identities = 48/131 (36%), Positives = 56/131 (42%), Gaps = 7/131 (5%)
Frame = +3
Query: 341 CLV*EVASFWGHSF*KWEFLRR*Q----VLHQHFPCFFHPPCQ*CNITI*IASQCLTLHT 396
C *E W F* E L + Q +L H P C N T * Q L L T
Sbjct: 1416 CWD*EADLLWVRFF*S*ESLLKCQAPQPLLQ*HSPRL----CLLWNTTC*NDFQFLMLFT 1583
Query: 397 LFW---LQP*LHLLASMW*EG*LQSLVVHQLLSSY*HPPFS*VQSV*VE*EFRT*L*SWK 453
W LQP L ++ *EG* L H S +* + VQS *VE F+
Sbjct: 1584 *AWWLLLQP---WLDNIL*EG*SSYLEEHHSSSLF*PAQYLSVQSH*VELAFQIWCTRLP 1754
Query: 454 IMSIWGLKIFA 464
IMSIW L+I A
Sbjct: 1755 IMSIWDLRISA 1787
>TC88574 weakly similar to GP|22137236|gb|AAM91463.1 At2g36630/F1O11.26
{Arabidopsis thaliana}, partial (25%)
Length = 663
Score = 218 bits (556), Expect = 3e-57
Identities = 106/203 (52%), Positives = 151/203 (74%)
Frame = +3
Query: 13 VIAATWLIMCILVMICNVSLAERVLKEKEPAKFVEKETKGFLKAMVDFLWESGKSSYEPV 72
V+ +++++ L++ C A R +K +E + V + + + E V
Sbjct: 96 VVLFSFILLFALLIFC----ANRTIKSEERSWIVNEAQSFYYHVKAN----------EHV 233
Query: 73 WPEMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIMG 132
WP+++ W+IIVG++IG GA+ GSVGGVGGGGIFVPML LIIGFD KS+TAISKCM+ G
Sbjct: 234 WPDIRLGWQIIVGTLIGIFGASFGSVGGVGGGGIFVPMLILIIGFDAKSATAISKCMVTG 413
Query: 133 AALSTVYYNMRLRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIIL 192
AA+STV++N++LR+PTLD+P+IDYDL LL P+++LGISIGV+ +V+FADWM+TVLLII+
Sbjct: 414 AAISTVFFNLKLRHPTLDIPMIDYDLVLLNAPVIILGISIGVVLSVVFADWMITVLLIIV 593
Query: 193 FIGTSTKALIKGINTWKKETMLK 215
FI TS +A +KG++TW KET++K
Sbjct: 594 FIVTSVRAYLKGLDTWNKETIIK 662
>TC77654 similar to GP|22137236|gb|AAM91463.1 At2g36630/F1O11.26
{Arabidopsis thaliana}, partial (94%)
Length = 1939
Score = 216 bits (550), Expect = 1e-56
Identities = 98/151 (64%), Positives = 129/151 (84%)
Frame = +2
Query: 70 EPVWPEMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCM 129
+ WPE+K W++ + SIIGFLG+A G+VGGVGGGGIFVPML LIIGFD KS+ A+SKCM
Sbjct: 212 DKTWPELKPSWRLALASIIGFLGSAFGTVGGVGGGGIFVPMLTLIIGFDTKSAAALSKCM 391
Query: 130 IMGAALSTVYYNMRLRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLL 189
IMGA+ S+V+YN+R+R+PT D+P++DYDLALLFQPMLMLGI++GV +V+F W++TVL+
Sbjct: 392 IMGASTSSVFYNLRVRHPTKDVPILDYDLALLFQPMLMLGITVGVALSVVFPYWLITVLI 571
Query: 190 IILFIGTSTKALIKGINTWKKETMLKKETAK 220
IILFIGTS+++ KG WK+ET+LKKE A+
Sbjct: 572 IILFIGTSSRSFFKGTEMWKEETLLKKEMAQ 664
Score = 34.3 bits (77), Expect = 0.10
Identities = 24/77 (31%), Positives = 41/77 (53%), Gaps = 4/77 (5%)
Frame = +2
Query: 75 EMKFDWK---IIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISK-CMI 130
E +W I+ ++ G LG +G + G GGG I P+L + IG P+ ++A + M+
Sbjct: 989 EASIEWTVLHIVFCALCGILGGTVGGLLGSGGGFILGPLL-IEIGVIPQVASATATFVMM 1165
Query: 131 MGAALSTVYYNMRLRNP 147
++LS V + + R P
Sbjct: 1166 FSSSLSVVEFYLLKRFP 1216
Score = 29.3 bits (64), Expect = 3.4
Identities = 26/79 (32%), Positives = 34/79 (42%), Gaps = 4/79 (5%)
Frame = +3
Query: 326 CYIVQLE**LVWLEGCLV*EVASFWGHSF*KWEFLRR*QVLHQHFPCFFHPPCQ*CNITI 385
C++ +E *+ LEG LV EV SFW + + V +H FH C N T
Sbjct: 1023 CFVRSVES*VGPLEGYLVLEVDSFWALFSLRSVSSLKLLVQQRHL**CFHHHCLWLNSTY 1202
Query: 386 *IAS----QCLTLHTLFWL 400
S C + LFWL
Sbjct: 1203 SRGSPFPMHCTSSQCLFWL 1259
>TC88695 similar to GP|13272465|gb|AAK17171.1 unknown protein {Arabidopsis
thaliana}, partial (24%)
Length = 591
Score = 100 bits (250), Expect = 9e-22
Identities = 46/125 (36%), Positives = 82/125 (64%), Gaps = 1/125 (0%)
Frame = +2
Query: 75 EMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIMGAA 134
+++ ++V I+ F+ +++ S GG+GGGGIF+P+L ++ G D K +++IS M+ G +
Sbjct: 218 QLQISVPLVVAGILCFIASSISSAGGIGGGGIFIPILTIVAGLDLKVASSISAFMVTGGS 397
Query: 135 LSTVY-YNMRLRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIILF 193
++ V Y LIDYD+AL +P ++LG+S+GVICN++F +W++T L+ +F
Sbjct: 398 IANVICYMFTTSTKFGGKSLIDYDIALSSEPCMLLGVSVGVICNLVFPEWLIT-LMFAVF 574
Query: 194 IGTST 198
+ ST
Sbjct: 575 LAWST 589
>TC82901 similar to GP|22137236|gb|AAM91463.1 At2g36630/F1O11.26
{Arabidopsis thaliana}, partial (22%)
Length = 647
Score = 75.5 bits (184), Expect = 4e-14
Identities = 46/129 (35%), Positives = 64/129 (48%), Gaps = 23/129 (17%)
Frame = +2
Query: 75 EMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGF----------------- 117
E+K W++ + SIIGFLG+A G+VGGVGGGGIFVPML LIIGF
Sbjct: 320 ELKPSWRLALASIIGFLGSAFGTVGGVGGGGIFVPMLTLIIGFRYQVCCCSFQMYDNGSI 499
Query: 118 ------DPKSSTAISKCMIMGAALSTVYYNMRLRNPTLDMPLIDYDLALLFQPMLMLGIS 171
PKS+++ +C +G + ++N+ MLGI+
Sbjct: 500 NIFCILQPKSASSNKRCSNIGFMI*LFFFNL----------------------CFMLGIT 613
Query: 172 IGVICNVMF 180
+GV NV+F
Sbjct: 614 VGVALNVVF 640
Score = 63.9 bits (154), Expect = 1e-10
Identities = 30/75 (40%), Positives = 51/75 (68%)
Frame = +3
Query: 109 PMLALIIGFDPKSSTAISKCMIMGAALSTVYYNMRLRNPTLDMPLIDYDLALLFQPMLML 168
P L L + D KS+ A+SKCMIMGA+ S+V+YN+R+R+PT D+P++D + F +
Sbjct: 423 PCLLLSLVLDTKSAAALSKCMIMGASTSSVFYNLRVRHPTKDVPILDL*SSSSFSTYALC 602
Query: 169 GISIGVICNVMFADW 183
+S+ V+ ++F+ +
Sbjct: 603 LVSLLVLLLMLFSPY 647
>CA920754 similar to PIR|T35781|T35 hypothetical protein SC8A6.14c SC8A6.14c
- Streptomyces coelicolor, partial (2%)
Length = 608
Score = 35.0 bits (79), Expect = 0.061
Identities = 25/74 (33%), Positives = 35/74 (46%), Gaps = 5/74 (6%)
Frame = -1
Query: 98 VGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIMGAALSTVYYNMR-----LRNPTLDMP 152
V GGGG+ P+ A S+TA S M+M V Y + ++NP L+ P
Sbjct: 443 VSAGGGGGVIFPVSASAAAAVSCSNTATSAAMMMDDLNRLVSYQQQQYCYNVQNPHLNHP 264
Query: 153 LIDYDLALLFQPML 166
++ LL QPML
Sbjct: 263 NPNHLSTLLMQPML 222
>TC91496 similar to GP|13272465|gb|AAK17171.1 unknown protein {Arabidopsis
thaliana}, partial (31%)
Length = 672
Score = 31.6 bits (70), Expect = 0.68
Identities = 18/59 (30%), Positives = 32/59 (53%), Gaps = 1/59 (1%)
Frame = +3
Query: 83 IVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMI-MGAALSTVYY 140
+V ++ L LG V G+GGG + P+L L +G P+ + A M+ + +S++ Y
Sbjct: 153 LVFPLMALLAGMLGGVFGIGGGMLISPLL-LQVGIAPEVTAATCSFMVFFSSTMSSLQY 326
>TC86391 similar to GP|4097571|gb|AAD09514.1| GMFP5 {Glycine max}, partial
(54%)
Length = 1353
Score = 30.8 bits (68), Expect = 1.2
Identities = 17/46 (36%), Positives = 24/46 (51%)
Frame = -3
Query: 148 TLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIILF 193
TL PL+ L+LL P L LG ++ N + + TVL +LF
Sbjct: 802 TLSNPLVPLFLSLLLNPCLHLGSPFQLLLNFIIILFSSTVLFFLLF 665
>AW585119
Length = 515
Score = 30.8 bits (68), Expect = 1.2
Identities = 16/45 (35%), Positives = 26/45 (57%), Gaps = 1/45 (2%)
Frame = -3
Query: 108 VPMLALIIGFD-PKSSTAISKCMIMGAALSTVYYNMRLRNPTLDM 151
+PM+ ++G PK+S AI C+I+ TV + L + TLD+
Sbjct: 336 IPMVVFLVGSQYPKTSVAIR*CLIISGFRGTVSVMVPLSHSTLDL 202
>TC82843 similar to PIR|A44226|A44226 auxin-independent growth promoter -
common tobacco, partial (56%)
Length = 1096
Score = 30.4 bits (67), Expect = 1.5
Identities = 18/62 (29%), Positives = 32/62 (51%), Gaps = 2/62 (3%)
Frame = -2
Query: 9 PSSLVIAATWLIMCILVMICNVSLAERVLKEKEPAKFVEKE--TKGFLKAMVDFLWESGK 66
P L+ A L M L+++ N SL+ + +K + + +E + L AMV F W+ G
Sbjct: 822 PKYLLRPANILAMLPLLLVTNTSLSSQTIKSRAAMREENEENGSSSSLVAMVSFEWKLGN 643
Query: 67 SS 68
++
Sbjct: 642 NA 637
>BF646944 similar to GP|12597808|gb| hypothetical protein {Arabidopsis
thaliana}, partial (14%)
Length = 340
Score = 29.6 bits (65), Expect = 2.6
Identities = 12/32 (37%), Positives = 21/32 (65%)
Frame = -2
Query: 178 VMFADWMVTVLLIILFIGTSTKALIKGINTWK 209
++F W+V V+ I++FI TS +L+ TW+
Sbjct: 156 IVFERWVVVVVGIVVFIDTS*CSLVTPFQTWE 61
>TC88533 similar to GP|12081919|dbj|BAB20862. plastidic cysteine synthase 1
{Solanum tuberosum}, partial (65%)
Length = 1136
Score = 28.5 bits (62), Expect = 5.7
Identities = 36/131 (27%), Positives = 55/131 (41%), Gaps = 18/131 (13%)
Frame = +1
Query: 14 IAATWLIMCILVMICNVSLAERVLKEKEPAKFVEKETKGFLKAMVDFLWESGKSSYE--- 70
IAA+ IL M ++SL RVL + A+ V E + V E KS+ +
Sbjct: 70 IAASKGYKLILTMPASMSLERRVLLKAFGAELVLTEAAKGMNGAVQKAEEIVKSTPDAYM 249
Query: 71 ------PVWPEMKFD------WKIIVGSIIGFLGAALGSVGGVGGGGIFVPM---LALII 115
P P++ F+ W+ G I L A +G+ G + G G F+ +I
Sbjct: 250 LQQFDNPSNPKIHFETTGPEIWEDTRGK-IDILVAGIGTGGTISGTGRFLKQQNSKVQVI 426
Query: 116 GFDPKSSTAIS 126
G +P S +S
Sbjct: 427 GVEPLESNILS 459
>TC89581 similar to PIR|B96661|B96661 unknown protein 83181-85105
[imported] - Arabidopsis thaliana, partial (35%)
Length = 716
Score = 28.1 bits (61), Expect = 7.5
Identities = 11/26 (42%), Positives = 17/26 (65%)
Frame = -3
Query: 199 KALIKGINTWKKETMLKKETAKQLEE 224
K L+ G+N WKK+ M E K++E+
Sbjct: 258 KVLLYGLNFWKKQLMKVNEFGKRIEK 181
>TC85727 homologue to PIR|D84431|D84431 probable endosomal protein
[imported] - Arabidopsis thaliana, partial (89%)
Length = 1925
Score = 27.7 bits (60), Expect = 9.8
Identities = 43/174 (24%), Positives = 69/174 (38%), Gaps = 21/174 (12%)
Frame = +3
Query: 80 WKIIVGSIIGF------LGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIM-- 131
WK I G + F AALGS + IF+ MLAL+ F P + A+ +++
Sbjct: 708 WKYIHGDVFRFPKFKSIFAAALGSGTQLFTLTIFIFMLALVGVFYPYNRGALFTALVVIY 887
Query: 132 -------GAALSTVYYNMR----LRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMF 180
G ++ Y + +RN L L L L+F + + I+ + F
Sbjct: 888 ALTSGIAGYTATSFYIQLEGTNWVRNLLLTGCLFCGPLFLMFCFLNTVAIAYSATAALPF 1067
Query: 181 ADWMVTVLLIILFIGTSTKALIKGI--NTWKKETMLKKETAKQLEEEPKTGAYQ 232
+ V+++I + TS ++ GI K E T K E P Y+
Sbjct: 1068G--TICVIVLIWTLVTSPLLVLGGIAGKNSKTEFQAPVRTTKYPREIPPLPWYR 1223
>TC80319 similar to GP|20259261|gb|AAM14366.1 putative HhoA protease
precursor {Arabidopsis thaliana}, partial (47%)
Length = 1195
Score = 27.7 bits (60), Expect = 9.8
Identities = 14/33 (42%), Positives = 18/33 (54%)
Frame = +1
Query: 114 IIGFDPKSSTAISKCMIMGAALSTVYYNMRLRN 146
IIGFDP A+ K + G L V Y + L+N
Sbjct: 598 IIGFDPSYDLAVLKVDVDGYELKPVGYLVNLKN 696
>CA920821 similar to GP|16930441|gb| At2g42810/F7D19.19 {Arabidopsis
thaliana}, partial (15%)
Length = 809
Score = 27.7 bits (60), Expect = 9.8
Identities = 13/37 (35%), Positives = 21/37 (56%)
Frame = -2
Query: 158 LALLFQPMLMLGISIGVICNVMFADWMVTVLLIILFI 194
L LL PML+ + G V+ W V +L+++LF+
Sbjct: 133 LGLLSFPMLLFVFAFGCFIFVL*WSWTVILLVLVLFV 23
>TC84247 weakly similar to GP|14269077|gb|AAK58011.1 verticillium wilt
disease resistance protein Ve2 {Lycopersicon
esculentum}, partial (7%)
Length = 836
Score = 27.7 bits (60), Expect = 9.8
Identities = 16/36 (44%), Positives = 20/36 (55%)
Frame = +2
Query: 76 MKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFVPML 111
MKFDW+ V + IGF GVG G +F PM+
Sbjct: 314 MKFDWQY-VSTGIGF---------GVGAGVVFAPMM 391
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.351 0.156 0.553
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,363,831
Number of Sequences: 36976
Number of extensions: 287442
Number of successful extensions: 2711
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 1996
Number of HSP's successfully gapped in prelim test: 85
Number of HSP's that attempted gapping in prelim test: 663
Number of HSP's gapped (non-prelim): 2145
length of query: 465
length of database: 9,014,727
effective HSP length: 100
effective length of query: 365
effective length of database: 5,317,127
effective search space: 1940751355
effective search space used: 1940751355
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147178.14


